Identification of a novel arsenite oxidase gene, arxA, in the haloalkaliphilic, arsenite-oxidizing bacterium Alkalilimnicola ehrlichii strain MLHE-1
- PMID: 20453090
- PMCID: PMC2897359
- DOI: 10.1128/JB.00244-10
Identification of a novel arsenite oxidase gene, arxA, in the haloalkaliphilic, arsenite-oxidizing bacterium Alkalilimnicola ehrlichii strain MLHE-1
Abstract
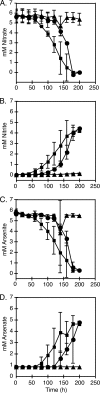
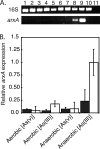
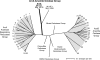
Although arsenic is highly toxic to most organisms, certain prokaryotes are known to grow on and respire toxic metalloids of arsenic (i.e., arsenate and arsenite). Two enzymes are known to be required for this arsenic-based metabolism: (i) the arsenate respiratory reductase (ArrA) and (ii) arsenite oxidase (AoxB). Both catalytic enzymes contain molybdopterin cofactors and form distinct phylogenetic clades (ArrA and AoxB) within the dimethyl sulfoxide (DMSO) reductase family of enzymes. Here we report on the genetic identification of a "new" type of arsenite oxidase that fills a phylogenetic gap between the ArrA and AoxB clades of arsenic metabolic enzymes. This "new" arsenite oxidase is referred to as ArxA and was identified in the genome sequence of the Mono Lake isolate Alkalilimnicola ehrlichii MLHE-1, a chemolithoautotroph that can couple arsenite oxidation to nitrate reduction. A genetic system was developed for MLHE-1 and used to show that arxA (gene locus ID mlg_0216) was required for chemoautotrophic arsenite oxidation. Transcription analysis also showed that mlg_0216 was only expressed under anaerobic conditions in the presence of arsenite. The mlg_0216 gene is referred to as arxA because of its greater homology to arrA relative to aoxB and previous reports that implicated Mlg_0216 (ArxA) of MLHE-1 in reversible arsenite oxidation and arsenate reduction in vitro. Our results and past observations support the position that ArxA is a distinct clade within the DMSO reductase family of proteins. These results raise further questions about the evolutionary relationships between arsenite oxidases (AoxB) and arsenate respiratory reductases (ArrA).
Figures


References
-
- Ahmann, D., A. L. Roberts, L. R. Krumholz, and F. M. Morel. 1994. Microbe grows by reducing arsenic. Nature 371:750. - PubMed
-
- Gralnick, J. A., C. T. Brown, and D. K. Newman. 2005. Anaerobic regulation by an atypical Arc system in Shewanella oneidensis. Mol. Microbiol. 56:1347-1357. - PubMed
-
- Hodak, H., A. Wohlkonig, C. Smet-Nocca, H. Drobecq, J. M. Wieruszeski, M. Senechal, I. Landrieu, C. Locht, M. Jamin, and F. Jacob-Dubuisson. 2008. The peptidyl-prolyl isomerase and chaperone Par27 of Bordetella pertussis as the prototype for a new group of parvulins. J. Mol. Biol. 376:414-426. - PubMed
-
- Hoeft, S. E., J. S. Blum, J. F. Stolz, F. R. Tabita, B. Witte, G. M. King, J. M. Santini, and R. S. Oremland. 2007. Alkalilimnicola ehrlichii sp nov., a novel, arsenite-oxidizing haloalkaliphilic gammaproteobacterium capable of chemoautotrophic or heterotrophic growth with nitrate or oxygen as the electron acceptor. Int. J. Syst. Evol. Microbiol. 57:504-512. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

