The hyperthermophilic euryarchaeon Archaeoglobus fulgidus repairs uracil by single-nucleotide replacement
- PMID: 20453094
- PMCID: PMC2953685
- DOI: 10.1128/JB.00135-10
The hyperthermophilic euryarchaeon Archaeoglobus fulgidus repairs uracil by single-nucleotide replacement
Abstract
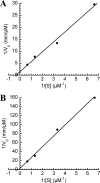
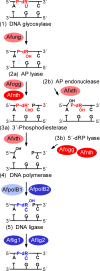
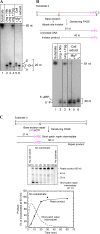
Hydrolytic deamination of cytosine to uracil in cellular DNA is a major source of C-to-T transition mutations if uracil is not repaired by the DNA base excision repair (BER) pathway. Since deamination increases rapidly with temperature, hyperthermophiles, in particular, are expected to succumb to such damage. There has been only one report of crenarchaeotic BER showing strong similarities to that in most eukaryotes and bacteria for hyperthermophilic Archaea. Here we report a different type of BER performed by extract prepared from cells of the euryarchaeon Archaeoglobus fulgidus. Although immunodepletion showed that the monofunctional family 4 type of uracil-DNA glycosylase (UDG) is the principal and probably only UDG in this organism, a β-elimination mechanism rather than a hydrolytic mechanism is employed for incision of the abasic site following uracil removal. The resulting 3' remnant is removed by efficient 3'-phosphodiesterase activity followed by single-nucleotide insertion and ligation. The finding that repair product formation is stimulated similarly by ATP and ADP in vitro raises the question of whether ADP is more important in vivo because of its higher heat stability.
Figures






References
-
- Akbari, M., M. Otterlei, J. Peña-Diaz, P. A. Aas, B. Kavli, N. B. Liabakk, L. Hagen, K. Imai, A. Durandy, G. Slupphaug, and H. E. Krokan. 2004. Repair of U/G and U/A in DNA by UNG2-associated repair complexes takes place predominantly by short-patch repair both in proliferating and growth-arrested cells. Nucleic Acids Res. 32:5486-5498. - PMC - PubMed
-
- Andersen, S., T. Heine, R. Sneve, I. König, H. E. Krokan, B. Epe, and H. Nilsen. 2005. Incorporation of dUMP into DNA is a major source of spontaneous DNA damage, while excision of uracil is not required for cytotoxicity of fluoropyrimidines in mouse embryonic fibroblasts. Carcinogenesis 26:547-555. - PubMed
-
- Birkeland, N.-K., H. Ånensen, I. Knævelsrud, W. Kristoffersen, M. Bjørås, F. T. Robb, A. Klungland, and S. Bjelland. 2002. Methylpurine DNA glycosylase of the hyperthermophilic archaeon Archaeoglobus fulgidus. Biochemistry 41:12697-12705. - PubMed
-
- Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

