Mutations in the Bacillus subtilis beta clamp that separate its roles in DNA replication from mismatch repair
- PMID: 20453097
- PMCID: PMC2897676
- DOI: 10.1128/JB.01435-09
Mutations in the Bacillus subtilis beta clamp that separate its roles in DNA replication from mismatch repair
Abstract
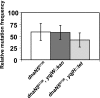
The beta clamp is an essential replication sliding clamp required for processive DNA synthesis. The beta clamp is also critical for several additional aspects of DNA metabolism, including DNA mismatch repair (MMR). The dnaN5 allele of Bacillus subtilis encodes a mutant form of beta clamp containing the G73R substitution. Cells with the dnaN5 allele are temperature sensitive for growth due to a defect in DNA replication at 49 degrees C, and they show an increase in mutation frequency caused by a partial defect in MMR at permissive temperatures. We selected for intragenic suppressors of dnaN5 that rescued viability at 49 degrees C to determine if the DNA replication defect could be separated from the MMR defect. We isolated three intragenic suppressors of dnaN5 that restored growth at the nonpermissive temperature while maintaining an increase in mutation frequency. All three dnaN alleles encoded the G73R substitution along with one of three novel missense mutations. The missense mutations isolated were S22P, S181G, and E346K. Of these, S181G and E346K are located near the hydrophobic cleft of the beta clamp, a common site occupied by proteins that bind the beta clamp. Using several methods, we show that the increase in mutation frequency resulting from each dnaN allele is linked to a defect in MMR. Moreover, we found that S181G and E346K allowed growth at elevated temperatures and did not have an appreciable effect on mutation frequency when separated from G73R. Thus, we found that specific residue changes in the B. subtilis beta clamp separate the role of the beta clamp in DNA replication from its role in MMR.
Figures




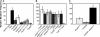


Similar articles
-
Binding of the regulatory domain of MutL to the sliding β-clamp is species specific.Nucleic Acids Res. 2019 May 21;47(9):4831-4842. doi: 10.1093/nar/gkz115. Nucleic Acids Res. 2019. PMID: 30916336 Free PMC article.
-
DnaN clamp zones provide a platform for spatiotemporal coupling of mismatch detection to DNA replication.Mol Microbiol. 2013 Feb;87(3):553-68. doi: 10.1111/mmi.12115. Epub 2012 Dec 11. Mol Microbiol. 2013. PMID: 23228104 Free PMC article.
-
Beta clamp directs localization of mismatch repair in Bacillus subtilis.Mol Cell. 2008 Feb 15;29(3):291-301. doi: 10.1016/j.molcel.2007.10.036. Mol Cell. 2008. PMID: 18280235 Free PMC article.
-
Mismatch Repair: From Preserving Genome Stability to Enabling Mutation Studies in Real-Time Single Cells.Cells. 2021 Jun 18;10(6):1535. doi: 10.3390/cells10061535. Cells. 2021. PMID: 34207040 Free PMC article. Review.
-
Mismatch repair in Gram-positive bacteria.Res Microbiol. 2016 Jan;167(1):4-12. doi: 10.1016/j.resmic.2015.08.006. Epub 2015 Sep 3. Res Microbiol. 2016. PMID: 26343983 Review.
Cited by
-
RecO and RecR are necessary for RecA loading in response to DNA damage and replication fork stress.J Bacteriol. 2014 Aug;196(15):2851-60. doi: 10.1128/JB.01494-14. Epub 2014 Jun 2. J Bacteriol. 2014. PMID: 24891441 Free PMC article.
-
Binding of the regulatory domain of MutL to the sliding β-clamp is species specific.Nucleic Acids Res. 2019 May 21;47(9):4831-4842. doi: 10.1093/nar/gkz115. Nucleic Acids Res. 2019. PMID: 30916336 Free PMC article.
-
DNA repair and genome maintenance in Bacillus subtilis.Microbiol Mol Biol Rev. 2012 Sep;76(3):530-64. doi: 10.1128/MMBR.05020-11. Microbiol Mol Biol Rev. 2012. PMID: 22933559 Free PMC article. Review.
-
Cost of rNTP/dNTP pool imbalance at the replication fork.Proc Natl Acad Sci U S A. 2013 Aug 6;110(32):12942-7. doi: 10.1073/pnas.1309506110. Epub 2013 Jul 23. Proc Natl Acad Sci U S A. 2013. PMID: 23882084 Free PMC article.
-
DnaN clamp zones provide a platform for spatiotemporal coupling of mismatch detection to DNA replication.Mol Microbiol. 2013 Feb;87(3):553-68. doi: 10.1111/mmi.12115. Epub 2012 Dec 11. Mol Microbiol. 2013. PMID: 23228104 Free PMC article.
References
-
- Berkmen, M. B., and A. D. Grossman. 2006. Spatial and temporal organization of the Bacillus subtilis replication cycle. Mol. Microbiol. 62:57-71. - PubMed
-
- Berkmen, M. B., and A. D. Grossman. 2007. Subcellular positioning of the origin region of the Bacillus subtilis chromosome is independent of sequences within oriC, the site of replication initiation, and the replication initiator DnaA. Mol. Microbiol. 63:150-165. - PubMed
-
- Beuning, P. J., D. Sawicka, D. Barsky, and G. C. Walker. 2006. Two processivity clamp interactions differentially alter the dual activities of UmuC. Mol. Microbiol. 59:460-474. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

