Disruption of the glycine cleavage system enables Sinorhizobium fredii USDA257 to form nitrogen-fixing nodules on agronomically improved North American soybean cultivars
- PMID: 20453144
- PMCID: PMC2897462
- DOI: 10.1128/AEM.00437-10
Disruption of the glycine cleavage system enables Sinorhizobium fredii USDA257 to form nitrogen-fixing nodules on agronomically improved North American soybean cultivars
Abstract
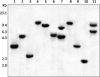
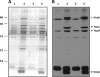
The symbiosis between Sinorhizobium fredii USDA257 and soybean [Glycine max (L.) Merr.] exhibits a high degree of cultivar specificity. USDA257 nodulates primitive soybean cultivars but fails to nodulate agronomically improved cultivars such as McCall. In this study we provide evidence for the involvement of a new genetic locus that controls soybean cultivar specificity. This locus was identified in USDA257 by Tn5 transposon mutagenesis followed by nodulation screening on McCall soybean. We have cloned the region corresponding to the site of Tn5 insertion and found that it lies within a 1.5-kb EcoRI fragment. DNA sequence analysis of this fragment and an adjacent 4.4-kb region identified an operon made up of three open reading frames encoding proteins of deduced molecular masses of 41, 13, and 104 kDa, respectively. These proteins revealed significant amino acid homology to glycine cleavage (gcv) system T, H, and P proteins of Escherichia coli and other organisms. Southern blot analysis revealed the presence of similar sequences in diverse rhizobia. Measurement of beta-galactosidase activity of a USDA257 strain containing a transcriptional fusion of gcvT promoter sequences to the lacZ gene revealed that the USDA257 gcvTHP operon was inducible by glycine. Inactivation of either gcvT or gcvP of USDA257 enabled the mutant to nodulate several agronomically improved North American soybean cultivars. These nodules revealed anatomical features typical of determinate nodules, with numerous bacteroids within the infected cells. Unlike for the previously characterized soybean cultivar specificity locus nolBTUVW, inactivation of the gcv locus had no discernible effect on the secretion of nodulation outer proteins of USDA257.
Figures



Similar articles
-
Molecular cloning and characterization of a sym plasmid locus that regulates cultivar-specific nodulation of soybean by Rhizobium fredii USDA257.Mol Microbiol. 1993 Jul;9(1):17-29. doi: 10.1111/j.1365-2958.1993.tb01665.x. Mol Microbiol. 1993. PMID: 8412662
-
Molecular aspects of soybean cultivar-specific nodulation by Sinorhizobium fredii USDA257.Indian J Exp Biol. 2003 Oct;41(10):1114-23. Indian J Exp Biol. 2003. PMID: 15242277 Review.
-
Sinorhizobium fredii USDA257, a cultivar-specific soybean symbiont, carries two copies of y4yA and y4yB, two open reading frames that are located in a region that encodes the type III protein secretion system.Mol Plant Microbe Interact. 2000 Sep;13(9):1010-4. doi: 10.1094/MPMI.2000.13.9.1010. Mol Plant Microbe Interact. 2000. PMID: 10975657
-
The soybean cultivar specificity gene nolX is present, expressed in a nodD-dependent manner, and of symbiotic significance in cultivar-nonspecific strains of Rhizobium (Sinorhizobium) fredii.Microbiology (Reading). 1997 Apr;143 ( Pt 4):1381-1388. doi: 10.1099/00221287-143-4-1381. Microbiology (Reading). 1997. PMID: 9141700
-
Symbiotic properties and first analyses of the genomic sequence of the fast growing model strain Sinorhizobium fredii HH103 nodulating soybean.J Biotechnol. 2011 Aug 20;155(1):11-9. doi: 10.1016/j.jbiotec.2011.03.016. Epub 2011 Mar 30. J Biotechnol. 2011. PMID: 21458507 Review.
Cited by
-
The Absence of the N-acyl-homoserine-lactone Autoinducer Synthase Genes traI and ngrI Increases the Copy Number of the Symbiotic Plasmid in Sinorhizobium fredii NGR234.Front Microbiol. 2016 Nov 18;7:1858. doi: 10.3389/fmicb.2016.01858. eCollection 2016. Front Microbiol. 2016. PMID: 27917168 Free PMC article.
-
Identification of Bradyrhizobium elkanii Genes Involved in Incompatibility with Soybean Plants Carrying the Rj4 Allele.Appl Environ Microbiol. 2015 Oct;81(19):6710-7. doi: 10.1128/AEM.01942-15. Epub 2015 Jul 17. Appl Environ Microbiol. 2015. PMID: 26187957 Free PMC article.
-
Metabolic transcriptional analysis on copper tolerance in moderate thermophilic bioleaching microorganism Acidithiobacillus caldus.J Ind Microbiol Biotechnol. 2020 Jan;47(1):21-33. doi: 10.1007/s10295-019-02247-6. Epub 2019 Nov 22. J Ind Microbiol Biotechnol. 2020. PMID: 31758413
-
Glycine Cleavage System and cAMP Receptor Protein Co-Regulate CRISPR/cas3 Expression to Resist Bacteriophage.Viruses. 2020 Jan 13;12(1):90. doi: 10.3390/v12010090. Viruses. 2020. PMID: 31941083 Free PMC article.
-
Replicon-dependent differentiation of symbiosis-related genes in Sinorhizobium strains nodulating Glycine max.Appl Environ Microbiol. 2014 Feb;80(4):1245-55. doi: 10.1128/AEM.03037-13. Epub 2013 Dec 6. Appl Environ Microbiol. 2014. PMID: 24317084 Free PMC article.
References
-
- Amadou, C., G. Pascal, S. Mangenot, M. Glew, C. Bontemps, D. Capela, S. Carrère, S. Cruveiller, C. Dossat, A. Lajus, M. Marchetti, V. Poinsot, Z. Rouy, B. Servin, M. Saad, C. Schenowitz, V. Barbe, J. Batut, C. Médigue, and C. Masson-Boivin. 2008. Genome sequence of the β-rhizobium Cupriavidus taiwanensis and comparative genomics of rhizobia. Genome Res. 18:1472-1483. - PMC - PubMed
-
- Balatti, P. A., and S. G. Pueppke. 1992. Identification of North American soybean lines that form nitrogen-fixing nodules with Rhizobium fredii USDA257. Can. J. Plant Sci. 72:49-55.
-
- Bec-Ferte, M.-P., H. B. Krishnan, D. Prome, A. Savagnac, S. G. Pueppke, and J.-C. Prome. 1994. Structures of nodulation factors from the nitrogen-fixing soybean symbiont Rhizobium fredii USDA257. Biochemistry 33:11782-11788. - PubMed
-
- Bellato, C., H. B. Krishnan, T. Cubo, F. Temprano, and S. G. Pueppke. 1997. The soybean cultivar specificity gene nolX is present, expressed in a nodD-dependent manner, and of symbiotic significance in cultivar-nonspecific strains of Rhizobium (Sinorhizobium) fredii. Microbiology 143:1381-1388. - PubMed
-
- Chatterjee, A., P. A. Balatti, W. Gibbons, and S. G. Pueppke. 1990. Interaction of Rhizobium fredii USDA257 and nodulation mutants derived from it with the agronomically improved soybean cultivar McCall. Planta 180:301-311. - PubMed
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

