Metagenomic analysis of apple orchard soil reveals antibiotic resistance genes encoding predicted bifunctional proteins
- PMID: 20453147
- PMCID: PMC2897439
- DOI: 10.1128/AEM.01763-09
Metagenomic analysis of apple orchard soil reveals antibiotic resistance genes encoding predicted bifunctional proteins
Abstract
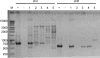
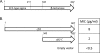
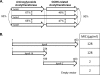
To gain insight into the diversity and origins of antibiotic resistance genes, we identified resistance genes in the soil in an apple orchard using functional metagenomics, which involves inserting large fragments of foreign DNA into Escherichia coli and assaying the resulting clones for expressed functions. Among 13 antibiotic-resistant clones, we found two genes that encode bifunctional proteins. One predicted bifunctional protein confers resistance to ceftazidime and contains a natural fusion between a predicted transcriptional regulator and a beta-lactamase. Sequence analysis of the entire metagenomic clone encoding the predicted bifunctional beta-lactamase revealed a gene potentially involved in chloramphenicol resistance as well as a predicted transposase. A second clone that encodes a predicted bifunctional protein confers resistance to kanamycin and contains an aminoglycoside acetyltransferase domain fused to a second acetyltransferase domain that, based on nucleotide sequence, was predicted not to be involved in antibiotic resistance. This is the first report of a transcriptional regulator fused to a beta-lactamase and of an aminoglycoside acetyltransferase fused to an acetyltransferase not involved in antibiotic resistance.
Figures
References
-
- Allen, H. K., L. A. Moe, J. Rodbumrer, A. Gaarder, and J. Handelsman. 2009. Functional metagenomics reveals diverse beta-lactamases in a remote Alaskan soil. ISME J. 3:243-251. - PubMed
-
- Ambler, R. P. 1980. The structure of beta-lactamases. Philos. Trans. R. Soc. Lond. B Biol. Sci. 289:321-331. - PubMed
-
- Aziz, R. K., D. Bartels, A. A. Best, M. DeJongh, T. Disz, R. A. Edwards, K. Formsma, S. Gerdes, E. M. Glass, M. Kubal, F. Meyer, G. J. Olsen, R. Olson, A. L. Osterman, R. A. Overbeek, L. K. McNeil, D. Paarmann, T. Paczian, B. Parrello, G. D. Pusch, C. Reich, R. Stevens, O. Vassieva, V. Vonstein, A. Wilke, and O. Zagnitko. 2008. The RAST server: rapid annotations using subsystems technology. BMC Genomics 9:75. - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources

