Cracking pre-40S ribosomal subunit structure by systematic analyses of RNA-protein cross-linking
- PMID: 20453830
- PMCID: PMC2892368
- DOI: 10.1038/emboj.2010.86
Cracking pre-40S ribosomal subunit structure by systematic analyses of RNA-protein cross-linking
Abstract
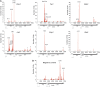
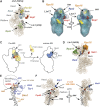
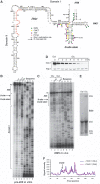
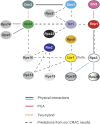
Understanding of eukaryotic ribosome synthesis has been slowed by a lack of structural data for the pre-ribosomal particles. We report rRNA-binding sites for six late-acting 40S ribosome synthesis factors, three of which cluster around the 3' end of the 18S rRNA in model 3D structures. Enp1 and Ltv1 were previously implicated in 'beak' structure formation during 40S maturation--and their binding sites indicate direct functions. The kinase Rio2, putative GTPase Tsr1 and dimethylase Dim1 bind sequences involved in tRNA interactions and mRNA decoding, indicating that their presence is incompatible with translation. The Dim1- and Tsr1-binding sites overlap with those of homologous Escherichia coli proteins, revealing conservation in assembly pathways. The primary binding sites for the 18S 3'-endonuclease Nob1 are distinct from its cleavage site and were unaltered by mutation of the catalytic PIN domain. Structure probing indicated that at steady state the cleavage site is likely unbound by Nob1 and flexible in the pre-rRNA. Nob1 binds before pre-rRNA cleavage, and we conclude that structural reorganization is needed to bring together the catalytic PIN domain and its target.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures

References
-
- Antunez de Mayolo P, Woolford JL Jr. (2003) Interactions of yeast ribosomal protein rpS14 with RNA. J Mol Biol 133: 697–709 - PubMed
-
- Ares M Jr, Igel AH (1990) Lethal and temperature-sensitive mutations and their suppressors identify an essential structural element in U2 small nuclear RNA. Genes Dev 4: 2132–2145 - PubMed
-
- Brodersen DE, Clemons WM Jr, Carter AP, Wimberly BT, Ramakrishnan V (2002) Crystal structure of the 30 S ribosomal subunit from Thermus thermophilus: structure of the proteins and their interactions with 16 S RNA. J Mol Biol 316: 725–768 - PubMed
-
- Brachmann CB, Davies A, Cost GJ, Caputo E, Li J, Hieter P, Boeke JD (1998) Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14: 115–132 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

