In vivo imaging of single-molecule translocation through nuclear pore complexes by pair correlation functions
- PMID: 20454622
- PMCID: PMC2862743
- DOI: 10.1371/journal.pone.0010475
In vivo imaging of single-molecule translocation through nuclear pore complexes by pair correlation functions
Abstract
Background: Nuclear pore complexes (NPCs) mediate bidirectional transport of proteins, RNAs, and ribonucleoproteins across the double-membrane nuclear envelope. Although there are many studies that look at the traffic in the nucleus and through the nuclear envelope we propose a method to detect the nucleocytoplasmic transport kinetics in an unperturbed cell, with no requirement for specific labeling of isolated molecules and, most important, in the presence of the cell milieu.
Methodology: The pair correlation function method (pCF) measures the time a molecule takes to migrate from one location to another within the cell in the presence of many molecules of the same kind. The spatial and temporal correlation among two arbitrary points in the cell provides a local map of molecular transport, and also highlights the presence of barriers to diffusion with millisecond time resolution and spatial resolution limited by diffraction. We use the pair correlation method to monitor a model protein substrate undergoing transport through NPCs in living cells, a biological problem in which single particle tracking (SPT) has given results that cannot be confirmed by traditional single-point FCS measurements because of the lack of spatial resolution.
Conclusions: We show that obstacles to molecular flow can be detected and that the pCF algorithm can recognize the heterogeneity of protein intra-compartment diffusion as well as the presence of barriers to transport across NE.
Conflict of interest statement
Figures

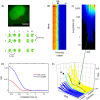
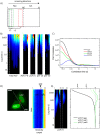



References
-
- Tran EJ, Wente SR. Dynamic nuclear pore complexes: life on the edge. Cell. 2006;125:1041–1053. - PubMed
-
- Gorlich D, Kutay U. Transport between the cell nucleus and the cytoplasm. Annu Rev Cell Dev Biol. 1999;15:607–660. - PubMed
-
- Rout MP, Aitchison JD. The nuclear pore complex as a transport machine. J Biol Chem. 2001;276:16593–16596. - PubMed

