Testing computational prediction of missense mutation phenotypes: functional characterization of 204 mutations of human cystathionine beta synthase
- PMID: 20455263
- PMCID: PMC3040297
- DOI: 10.1002/prot.22722
Testing computational prediction of missense mutation phenotypes: functional characterization of 204 mutations of human cystathionine beta synthase
Abstract
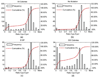
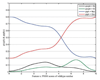
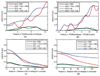
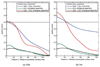
Predicting the phenotypes of missense mutations uncovered by large-scale sequencing projects is an important goal in computational biology. High-confidence predictions can be an aid in focusing experimental and association studies on those mutations most likely to be associated with causative relationships between mutation and disease. As an aid in developing these methods further, we have derived a set of random mutations of the enzymatic domains of human cystathionine beta synthase. This enzyme is a dimeric protein that catalyzes the condensation of serine and homocysteine to produce cystathionine. Yeast missing this enzyme cannot grow on medium lacking a source of cysteine, while transfection of functional human CBS into yeast strains missing endogenous enzyme can successfully complement for the missing gene. We used PCR mutagenesis with error-prone Taq polymerase to produce 948 colonies and compared cell growth in the presence or absence of a cysteine source as a measure of CBS function. We were able to infer the phenotypes of 204 single-site mutants, 79 of them deleterious and 125 neutral. This set was used to test the accuracy of six publicly available prediction methods for phenotype prediction of missense mutations: SIFT, PolyPhen, PMut, SNPs3D, PhD-SNP, and nsSNPAnalyzer. The top methods are PolyPhen, SIFT, and nsSNPAnalyzer, which have similar performance. Using kernel discriminant functions, we found that the difference in position-specific scoring matrix values is more predictive than the wild-type PSSM score alone, and that the relative surface area in the biologically relevant complex is more predictive than that of the monomeric proteins.
Figures

References
-
- Collins FS, Patrinos A, Jordan E, Chakravarti A, Gesteland R, Walters L. New goals for the U.S. human genome project: 1998–2003. Science. 1998;282:682–689. - PubMed
-
- Carlson KM, Dou S, Chi D, Scavarda N, Toshima K, Jackson CE, Wells SA, Jr, Goodfellow PJ, Donis-Keller H. Single missense mutation in the tyrosine kinase catalytic domain of the RET protooncogene is associated with multiple endocrine neoplasia type 2B. Proc Natl Acad Sci USA. 1994;91:1579–1583. - PMC - PubMed
-
- Garrigue-Antar L, Munoz-Antonia T, Antonia SJ, Gesmonde J, Vellucci VF, Reiss M. Missense mutations of the transforming growth factor beta type II receptor in human head and neck squamous carcinoma cells. Cancer Res. 1995;55:3982–3987. - PubMed
-
- Marchese CA, Bertolino F, Ceccopieri B, Vanzetti M, Scaglione D, Locatelli L, Montera M, Romio L, Resta N, Stella A, Guanti G, Mareni C. Clinical findings in a family with familial adenomatous polyposis and a missense mutation of the adenomatous polyposis coli gene. Scand J Gastroenterol. 1996;31:917–920. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

