Scaffold rearrangement of dihydroxypyrimidine inhibitors of HIV integrase: Docking model revisited
- PMID: 20457521
- PMCID: PMC7323870
- DOI: 10.1016/j.bmcl.2010.04.048
Scaffold rearrangement of dihydroxypyrimidine inhibitors of HIV integrase: Docking model revisited
Abstract
A series of dihydroxypyrimidine (DHP) derivatives were designed as inhibitors of HIV integrase (IN) based on known homology models. Through chemical synthesis and biochemical assays it was found that the activity profile of these compounds largely deviates from predictions with existing models. With the recently disclosed IN crystal structure of prototype foamy virus (PFV), a new HIV IN homology model was constructed featuring a critical IN/DNA interface previously lacking. With this new model, docking results completely corroborated observed biological activities. This new model should provide a more accurate and improved platform for the design of new inhibitors of HIV IN.
Copyright 2010 Elsevier Ltd. All rights reserved.
Figures
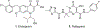




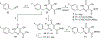

Similar articles
-
3-Hydroxypyrimidine-2,4-diones as an inhibitor scaffold of HIV integrase.J Med Chem. 2011 Apr 14;54(7):2282-92. doi: 10.1021/jm1014378. Epub 2011 Mar 7. J Med Chem. 2011. PMID: 21381765 Free PMC article.
-
Multivariate QSAR study of 4,5-dihydroxypyrimidine carboxamides as HIV-1 integrase inhibitors.Eur J Med Chem. 2009 Sep;44(9):3577-83. doi: 10.1016/j.ejmech.2009.03.001. Epub 2009 Mar 9. Eur J Med Chem. 2009. PMID: 19327872
-
Study on the interactions between diketo-acid inhibitors and prototype foamy virus integrase-DNA complex via molecular docking and comparative molecular dynamics simulation methods.J Biomol Struct Dyn. 2013;31(7):734-47. doi: 10.1080/07391102.2012.709458. Epub 2012 Aug 22. J Biomol Struct Dyn. 2013. PMID: 22913375
-
Dipyrimidine-based inhibitors of HIV-1 integrase.Expert Opin Investig Drugs. 2003 Feb;12(2):289-92. doi: 10.1517/13543784.12.2.289. Expert Opin Investig Drugs. 2003. PMID: 12556222 Review. No abstract available.
-
In search of second-generation HIV integrase inhibitors: targeting integration beyond strand transfer.Future Med Chem. 2009 Oct;1(7):1259-74. doi: 10.4155/fmc.09.86. Future Med Chem. 2009. PMID: 21426102 Review.
Cited by
-
4,5-Dihydroxypyrimidine Methyl Carboxylates, Carboxylic Acids, and Carboxamides as Inhibitors of Human Cytomegalovirus pUL89 Endonuclease.J Med Chem. 2022 Apr 14;65(7):5830-5849. doi: 10.1021/acs.jmedchem.2c00203. Epub 2022 Apr 4. J Med Chem. 2022. PMID: 35377638 Free PMC article.
-
3-Hydroxypyrimidine-2,4-diones as an inhibitor scaffold of HIV integrase.J Med Chem. 2011 Apr 14;54(7):2282-92. doi: 10.1021/jm1014378. Epub 2011 Mar 7. J Med Chem. 2011. PMID: 21381765 Free PMC article.
-
Molecular mechanisms of retroviral integrase inhibition and the evolution of viral resistance.Proc Natl Acad Sci U S A. 2010 Nov 16;107(46):20057-62. doi: 10.1073/pnas.1010246107. Epub 2010 Oct 28. Proc Natl Acad Sci U S A. 2010. PMID: 21030679 Free PMC article.
-
Basic quinolinonyl diketo acid derivatives as inhibitors of HIV integrase and their activity against RNase H function of reverse transcriptase.J Med Chem. 2014 Apr 24;57(8):3223-34. doi: 10.1021/jm5001503. Epub 2014 Apr 11. J Med Chem. 2014. PMID: 24684270 Free PMC article.
-
Structural biology of retroviral DNA integration.Virology. 2011 Mar 15;411(2):194-205. doi: 10.1016/j.virol.2010.12.008. Epub 2011 Jan 8. Virology. 2011. PMID: 21216426 Free PMC article. Review.
References
-
- Asante-Appiah E; Skalka AM Antiviral. Res. 1997, 36, 139. - PubMed
-
- Pommier Y; Johnson AA; Marchand C Nat. Rev. Drug Discovery 2005, 4, 236. - PubMed
-
- Coiras M; Lopez-Huertas MR; Perez-Olmeda M; Alcami J Nat. Rev. Microbiol. 2009, 7, 798. - PubMed
-
- Dahl V; Josefsson L; Palmer S Antiviral. Res. 2009, 85, 286. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

