Comparative genomics and evolution of the alpha-defensin multigene family in primates
- PMID: 20457584
- PMCID: PMC2981490
- DOI: 10.1093/molbev/msq118
Comparative genomics and evolution of the alpha-defensin multigene family in primates
Abstract
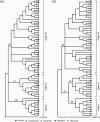
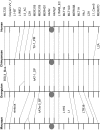
Defensin genes encode small cationic antimicrobial peptides that form an important part of the innate immune system. They are divided into three families, alpha (α), beta (β), and theta (), according to arrangement of the disulfide bonding pattern between cysteine residues. Considering the functional importance of defensins, investigators have studied the evolution and the genomic organization of defensin genes. However, these studies have been restricted mainly to β-defensins. To understand the evolutionary dynamics of α-defensin genes among primates, we identified the α-defensin repertoires in human, chimpanzee, orangutan, macaque, and marmoset. The α-defensin genes in primates can be classified into three phylogenetic classes (class I, II, and III). The presence of all three classes in the marmoset indicates that their divergence occurred before the separation of New World and Old World monkeys. Comparative analysis of the α-defensin genomic clusters suggests that the makeup of the α-defensin gene repertoires between primates is quite different, as their genes have undergone dramatic birth-and-death evolution. Analysis of the encoded peptides of the α-defensin genes indicates that despite the overall high level of sequence divergence, certain amino acid residues or motifs are conserved within and between the three phylogenetic classes. The evolution of α-defensins in primates, therefore, appears to be governed by two opposing evolutionary forces. One force stabilizes specific amino acid residues and motifs to preserve the functional and structural integrity of the molecules and the other diversifies the sequences generating molecules with a wide range of activities against a large number of pathogens.
Figures



Similar articles
-
Molecular evolution of the primate α-/θ-defensin multigene family.PLoS One. 2014 May 12;9(5):e97425. doi: 10.1371/journal.pone.0097425. eCollection 2014. PLoS One. 2014. PMID: 24819937 Free PMC article.
-
Rapid evolution and diversification of mammalian alpha-defensins as revealed by comparative analysis of rodent and primate genes.Physiol Genomics. 2004 Dec 15;20(1):1-11. doi: 10.1152/physiolgenomics.00150.2004. Epub 2004 Oct 19. Physiol Genomics. 2004. PMID: 15494476
-
Evolution of primate α and θ defensins revealed by analysis of genomes.Mol Biol Rep. 2014 Jun;41(6):3859-66. doi: 10.1007/s11033-014-3253-z. Epub 2014 Feb 21. Mol Biol Rep. 2014. PMID: 24557891
-
Rapid sequence divergence in mammalian beta-defensins by adaptive evolution.Mol Immunol. 2003 Nov;40(7):413-21. doi: 10.1016/s0161-5890(03)00160-3. Mol Immunol. 2003. PMID: 14568387 Review.
-
The changing of the guard: Molecular diversity and rapid evolution of beta-defensins.Mol Divers. 2006 Nov;10(4):575-84. doi: 10.1007/s11030-006-9031-7. Mol Divers. 2006. PMID: 16969721 Review.
Cited by
-
VP4 Is a Determinant of Alpha-Defensin Modulation of Rotaviral Infection.J Virol. 2022 Apr 13;96(7):e0205321. doi: 10.1128/jvi.02053-21. Epub 2022 Mar 14. J Virol. 2022. Corrected and republished in: J Virol. 2023 Oct 31;97(10):e0096223. doi: 10.1128/jvi.00962-23. PMID: 35285683 Free PMC article. Corrected and republished.
-
PvML1 suppresses bacterial infection by recognizing LPS and regulating AMP expression in shrimp.Front Immunol. 2022 Dec 28;13:1088862. doi: 10.3389/fimmu.2022.1088862. eCollection 2022. Front Immunol. 2022. PMID: 36643915 Free PMC article.
-
Enhancement of antiviral activity of human alpha-defensin 5 against herpes simplex virus 2 by arginine mutagenesis at adaptive evolution sites.J Virol. 2013 Mar;87(5):2835-45. doi: 10.1128/JVI.02209-12. Epub 2012 Dec 26. J Virol. 2013. PMID: 23269800 Free PMC article.
-
Abundant β-Defensin Copy Number Variations in Pigs.Genes (Basel). 2025 Apr 4;16(4):430. doi: 10.3390/genes16040430. Genes (Basel). 2025. PMID: 40282390 Free PMC article.
-
Performance evaluation of antimicrobial peptide ll-37 and hepcidin and β-defensin-2 secreted by mesenchymal stem cells.Heliyon. 2019 Oct 23;5(10):e02652. doi: 10.1016/j.heliyon.2019.e02652. eCollection 2019 Oct. Heliyon. 2019. PMID: 31687504 Free PMC article. Review.
References
-
- Arnold K, Bordoli L, Kopp J, Schwede T. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics. 2006;22:195–201. - PubMed
-
- Bennett-Lovsey RM, Herbert AD, Sternberg MJ, Kelley LA. Exploring the extremes of sequence/structure space with ensemble fold recognition in the program Phyre. Proteins. 2008;70:611–625. - PubMed
-
- Boniotto M, Tossi A, DelPero M, Sgubin S, Antcheva N, Santon D, Masters J, Crovella S. Evolution of the beta defensin 2 gene in primates. Genes Immun. 2003;4:251–257. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

