Tyrosine residues at the carboxyl terminus of Vav1 play an important role in regulation of its biological activity
- PMID: 20457609
- PMCID: PMC2906301
- DOI: 10.1074/jbc.M109.094508
Tyrosine residues at the carboxyl terminus of Vav1 play an important role in regulation of its biological activity
Abstract
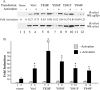
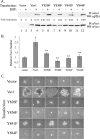
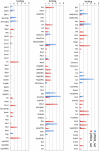
The guanine nucleotide exchange factor (GEF) Vav1 is an essential signal transducer protein in the hematopoietic system, where it is expressed physiologically. It is also involved in several human malignancies. Tyrosine phosphorylation at the Vav1 amino terminus plays a central role in regulating its activity; however, the role of carboxyl terminal tyrosine residues is unknown. We found that mutation of either Tyr-826 (Y826F) or Tyr-841 (Y841F) to phenylalanine led to loss of Vav1 GEF activity. When these Vav1 mutants were ectopically expressed in pancreatic cancer cells lacking Vav1, they failed to induce growth in agar, indicating loss of transforming potential. Furthermore, although Y841F had no effect on Vav1-stimulated nuclear factor of activated T cells (NFAT) activity, Y826F doubled NFAT activity when compared with Vav1, suggesting that Tyr-826 mediates an autoinhibitory effect on NFAT activity. SH2 profiling revealed that Shc, Csk, Abl, and Sap associate with Tyr-826, whereas SH2-B, Src, Brk, GTPase-activating protein, and phospholipase C-gamma associate with Tyr-841. Although the mutations in the Tyr-826 and Tyr-841 did not affect the binding of the carboxyl SH3 of Vav1 to other proteins, binding to several of the proteins identified by the SH2 profiling was lost. Of interest is Csk, which associates with wild-type Vav1 and Y841F, yet it fails to associate with Y826F, suggesting that loss of binding between Y826F and Csk might relieve an autoinhibitory effect, leading to increased NFAT. Our data indicate that GEF activity is critical for the function of Vav1 as a transforming protein but not for NFAT stimulation. The association of Vav1 with other proteins, detected by SH2 profiling, might affect other Vav1-dependent activities, such as NFAT stimulation.
Figures





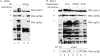
Similar articles
-
The association of Sam68 with Vav1 contributes to tumorigenesis.Cell Signal. 2007 Dec;19(12):2479-86. doi: 10.1016/j.cellsig.2007.07.022. Epub 2007 Aug 8. Cell Signal. 2007. PMID: 17855053
-
A guanine nucleotide exchange factor-independent function of Vav1 in transcriptional activation.J Biol Chem. 2000 Jan 21;275(3):2185-90. doi: 10.1074/jbc.275.3.2185. J Biol Chem. 2000. PMID: 10636924
-
Vav1 and Ly-GDI two regulators of Rho GTPases, function cooperatively as signal transducers in T cell antigen receptor-induced pathways.J Biol Chem. 2002 Dec 20;277(51):50121-30. doi: 10.1074/jbc.M204299200. Epub 2002 Oct 16. J Biol Chem. 2002. PMID: 12386169
-
Flesh and blood: the story of Vav1, a gene that signals in hematopoietic cells but can be transforming in human malignancies.Cancer Lett. 2007 Oct 8;255(2):241-54. doi: 10.1016/j.canlet.2007.04.015. Epub 2007 Jun 21. Cancer Lett. 2007. PMID: 17590270 Review.
-
Unraveling the Oncogenic Potential of VAV1 in Human Cancer: Lessons from Mouse Models.Cells. 2023 Apr 27;12(9):1276. doi: 10.3390/cells12091276. Cells. 2023. PMID: 37174676 Free PMC article. Review.
Cited by
-
Dynamin 2 potentiates invasive migration of pancreatic tumor cells through stabilization of the Rac1 GEF Vav1.Dev Cell. 2013 Mar 25;24(6):573-85. doi: 10.1016/j.devcel.2013.02.010. Dev Cell. 2013. PMID: 23537630 Free PMC article.
-
Structure analysis between the SWAP-70 RHO-GEF and the newly described PLD2-GEF.Small GTPases. 2012 Oct-Dec;3(4):202-8. doi: 10.4161/sgtp.20887. Epub 2012 Aug 3. Small GTPases. 2012. PMID: 22858691 Free PMC article.
-
Vav1 Regulates T-Cell Activation through a Feedback Mechanism and Crosstalk between the T-Cell Receptor and CD28.J Proteome Res. 2015 Jul 2;14(7):2963-75. doi: 10.1021/acs.jproteome.5b00340. Epub 2015 Jun 16. J Proteome Res. 2015. PMID: 26043137 Free PMC article.
-
Vav1 in hematologic neoplasms, a mini review.Am J Blood Res. 2012;2(1):1-8. Epub 2012 Jan 1. Am J Blood Res. 2012. PMID: 22432082 Free PMC article.
-
Proteomic and phosphoproteomic landscapes of acute myeloid leukemia.Blood. 2022 Sep 29;140(13):1533-1548. doi: 10.1182/blood.2022016033. Blood. 2022. PMID: 35895896 Free PMC article.
References
-
- Sjöblom T., Jones S., Wood L. D., Parsons D. W., Lin J., Barber T. D., Mandelker D., Leary R. J., Ptak J., Silliman N., Szabo S., Buckhaults P., Farrell C., Meeh P., Markowitz S. D., Willis J., Dawson D., Willson J. K., Gazdar A. F., Hartigan J., Wu L., Liu C., Parmigiani G., Park B. H., Bachman K. E., Papadopoulos N., Vogelstein B., Kinzler K. W., Velculescu V. E. (2006) Science 314, 268–274 - PubMed
-
- Hunter T. (2000) Cell 100, 113–127 - PubMed
-
- Schlessinger J. (2000) Cell 103, 211–225 - PubMed
-
- Manning G., Whyte D. B., Martinez R., Hunter T., Sudarsanam S. (2002) Science 298, 1912–1934 - PubMed
-
- Alonso A., Sasin J., Bottini N., Friedberg I., Friedberg I., Osterman A., Godzik A., Hunter T., Dixon J., Mustelin T. (2004) Cell 117, 699–711 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

