In vitro genetic reconstruction of bacterial transcription initiation by coupled synthesis and detection of RNA polymerase holoenzyme
- PMID: 20457746
- PMCID: PMC2910072
- DOI: 10.1093/nar/gkq377
In vitro genetic reconstruction of bacterial transcription initiation by coupled synthesis and detection of RNA polymerase holoenzyme
Abstract
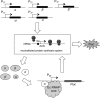
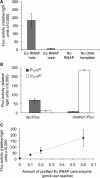
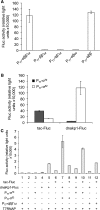
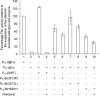
In vitro reconstitution of a biological complex or process normally involves assembly of multiple individually purified protein components. Here we present a strategy that couples expression and assembly of multiple gene products with functional detection in an in vitro reconstituted protein synthesis system. The strategy potentially allows experimental reconstruction of a multi-component biological complex or process using only DNA templates instead of purified proteins. We applied this strategy to bacterial transcription initiation by co-expressing genes encoding Escherichia coli RNA polymerase subunits and sigma factors in the reconstituted protein synthesis system and by coupling the synthesis and assembly of a functional RNA polymerase holoenzyme with the expression of a reporter gene. Using such a system, we demonstrated sigma-factor-dependent, promoter-specific transcription initiation. Since protein synthesis, complex formation and enzyme catalysis occur in the same in vitro reaction mixture, this reconstruction process resembles natural biosynthetic pathways and avoids time-consuming expression and purification of individual proteins. The strategy can significantly reduce the time normally required by conventional reconstitution methods, allow rapid generation and detection of genetic mutations, and provide an open and designable platform for in vitro study and intervention of complex biological processes.
Figures
References
-
- Yuzhakov A, Turner J, O'Donnell M. Replisome assembly reveals the basis for asymmetric function in leading and lagging strand replication. Cell. 1996;86:877–886. - PubMed
-
- Fujita N, Ishihama A. Reconstitution of RNA polymerase. Methods Enzymol. 1996;273:121–130. - PubMed
-
- Shimizu Y, Inoue A, Tomari Y, Suzuki T, Yokogawa T, Nishikawa K, Ueda T. Cell-free translation reconstituted with purified components. Nat. Biotechnol. 2001;19:751–755. - PubMed
-
- Kung HF, Redfield B, Treadwell BV, Eskin B, Spears C, Weissbach H. DNA-directed in vitro synthesis of beta-galactosidase. Studies with purified factors. J. Biol. Chem. 1977;252:6889–6894. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

