Metnase promotes restart and repair of stalled and collapsed replication forks
- PMID: 20457750
- PMCID: PMC2943610
- DOI: 10.1093/nar/gkq339
Metnase promotes restart and repair of stalled and collapsed replication forks
Abstract
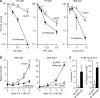
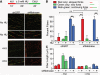
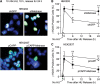
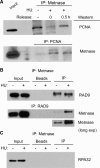
Metnase is a human protein with methylase (SET) and nuclease domains that is widely expressed, especially in proliferating tissues. Metnase promotes non-homologous end-joining (NHEJ), and knockdown causes mild hypersensitivity to ionizing radiation. Metnase also promotes plasmid and viral DNA integration, and topoisomerase IIα (TopoIIα)-dependent chromosome decatenation. NHEJ factors have been implicated in the replication stress response, and TopoIIα has been proposed to relax positive supercoils in front of replication forks. Here we show that Metnase promotes cell proliferation, but it does not alter cell cycle distributions, or replication fork progression. However, Metnase knockdown sensitizes cells to replication stress and confers a marked defect in restart of stalled replication forks. Metnase promotes resolution of phosphorylated histone H2AX, a marker of DNA double-strand breaks at collapsed forks, and it co-immunoprecipitates with PCNA and RAD9, a member of the PCNA-like RAD9-HUS1-RAD1 intra-S checkpoint complex. Metnase also promotes TopoIIα-mediated relaxation of positively supercoiled DNA. Metnase is not required for RAD51 focus formation after replication stress, but Metnase knockdown cells show increased RAD51 foci in the presence or absence of replication stress. These results establish Metnase as a key factor that promotes restart of stalled replication forks, and implicate Metnase in the repair of collapsed forks.
Figures
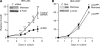
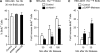

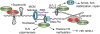
Similar articles
-
Chk1 phosphorylation of Metnase enhances DNA repair but inhibits replication fork restart.Oncogene. 2012 Sep 20;31(38):4245-54. doi: 10.1038/onc.2011.586. Epub 2012 Jan 9. Oncogene. 2012. PMID: 22231448 Free PMC article.
-
Metnase mediates chromosome decatenation in acute leukemia cells.Blood. 2009 Aug 27;114(9):1852-8. doi: 10.1182/blood-2008-08-175760. Epub 2009 May 20. Blood. 2009. PMID: 19458360 Free PMC article.
-
Metnase/SETMAR: a domesticated primate transposase that enhances DNA repair, replication, and decatenation.Genetica. 2010 May;138(5):559-66. doi: 10.1007/s10709-010-9452-1. Epub 2010 Mar 23. Genetica. 2010. PMID: 20309721 Free PMC article. Review.
-
Metnase Mediates Loading of Exonuclease 1 onto Single Strand Overhang DNA for End Resection at Stalled Replication Forks.J Biol Chem. 2017 Jan 27;292(4):1414-1425. doi: 10.1074/jbc.M116.745646. Epub 2016 Dec 14. J Biol Chem. 2017. PMID: 27974460 Free PMC article.
-
Metnase and EEPD1: DNA Repair Functions and Potential Targets in Cancer Therapy.Front Oncol. 2022 Jan 28;12:808757. doi: 10.3389/fonc.2022.808757. eCollection 2022. Front Oncol. 2022. PMID: 35155245 Free PMC article. Review.
Cited by
-
Histone modifications and DNA double-strand break repair after exposure to ionizing radiations.Radiat Res. 2013 Apr;179(4):383-92. doi: 10.1667/RR3308.2. Epub 2013 Feb 1. Radiat Res. 2013. PMID: 23373901 Free PMC article. Review.
-
Recovery from the DNA Replication Checkpoint.Genes (Basel). 2016 Oct 28;7(11):94. doi: 10.3390/genes7110094. Genes (Basel). 2016. PMID: 27801838 Free PMC article. Review.
-
SETMAR Shorter Isoform: A New Prognostic Factor in Glioblastoma.Front Oncol. 2022 Jan 3;11:638397. doi: 10.3389/fonc.2021.638397. eCollection 2021. Front Oncol. 2022. PMID: 35047379 Free PMC article.
-
WDR5 supports colon cancer cells by promoting methylation of H3K4 and suppressing DNA damage.BMC Cancer. 2018 Jun 20;18(1):673. doi: 10.1186/s12885-018-4580-6. BMC Cancer. 2018. PMID: 29925347 Free PMC article.
-
Deletion of Mouse Setd4 Promotes the Recovery of Hematopoietic Failure.Int J Radiat Oncol Biol Phys. 2020 Jul 15;107(4):779-792. doi: 10.1016/j.ijrobp.2020.03.026. Epub 2020 Apr 4. Int J Radiat Oncol Biol Phys. 2020. PMID: 32259569 Free PMC article.
References
-
- Hanks S, Coleman K, Reid S, Plaja A, Firth H, FitzPatrick D, Kidd A, Méhes K, Nash R, Robin N, et al. Constitutional aneuploidy and cancer predisposition caused by biallelic mutations in BUB1B. Nat. Genet. 2004;36:1159–1161. - PubMed
-
- Burma S, Chen BP, Chen DJ. Role of non-homologous end joining (NHEJ) in maintaining genomic integrity. DNA Repair. 2006;5:1042–1048. - PubMed
-
- Shimada K, Oma Y, Schleker T, Kugou K, Ohta K, Harata M, Gasser SM. Ino80 chromatin remodeling complex promotes recovery of stalled replication forks. Curr. Biol. 2008;18:566–575. - PubMed
-
- Budzowska M, Kanaar R. Mechanisms of dealing with DNA damage-induced replication problems. Cell Biochem. Biophys. 2009;53:17–31. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- T32 CA009582/CA/NCI NIH HHS/United States
- S10 RR14668/RR/NCRR NIH HHS/United States
- T32 CA09582/CA/NCI NIH HHS/United States
- R01 GM033944/GM/NIGMS NIH HHS/United States
- R01 GM084020/GM/NIGMS NIH HHS/United States
- R01GM084020/GM/NIGMS NIH HHS/United States
- S10 RR19287/RR/NCRR NIH HHS/United States
- P20 RR11830/RR/NCRR NIH HHS/United States
- F31 CA132628/CA/NCI NIH HHS/United States
- S10RR016918/RR/NCRR NIH HHS/United States
- R01 CA100862/CA/NCI NIH HHS/United States
- P30 CA118100/CA/NCI NIH HHS/United States
- R01 HL093606/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

