Pseudomonas aeruginosa evasion of phagocytosis is mediated by loss of swimming motility and is independent of flagellum expression
- PMID: 20457788
- PMCID: PMC2897393
- DOI: 10.1128/IAI.00144-10
Pseudomonas aeruginosa evasion of phagocytosis is mediated by loss of swimming motility and is independent of flagellum expression
Abstract
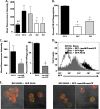
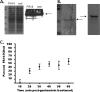
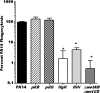
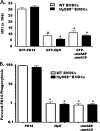
Pseudomonas aeruginosa is a pathogenic Gram-negative bacterium that causes severe opportunistic infections in immunocompromised individuals; in particular, severity of infection with P. aeruginosa positively correlates with poor prognosis in cystic fibrosis (CF) patients. Establishment of chronic infection by this pathogen is associated with downregulation of flagellar expression and of other genes that regulate P. aeruginosa motility. The current paradigm is that loss of flagellar expression enables immune evasion by the bacteria due to loss of engagement by phagocytic receptors that recognize flagellar components and loss of immune activation through flagellin-mediated Toll-like receptor (TLR) signaling. In this work, we employ bacterial and mammalian genetic approaches to demonstrate that loss of motility, not the loss of the flagellum per se, is the critical factor in the development of resistance to phagocytosis by P. aeruginosa. We demonstrate that isogenic P. aeruginosa mutants deficient in flagellar function, but retaining an intact flagellum, are highly resistant to phagocytosis by both murine and human phagocytic cells at levels comparable to those of flagellum-deficient mutants. Furthermore, we show that loss of MyD88 signaling in murine phagocytes does not recapitulate the phagocytic deficit observed for either flagellum-deficient or motility-deficient P. aeruginosa mutants. Our data demonstrate that loss of bacterial motility confers a dramatic resistance to phagocytosis that is independent of both flagellar expression and TLR signaling. These findings provide an explanation for the well-documented observation of nonmotility in clinical P. aeruginosa isolates and for how this phenotype confers upon the bacteria an advantage in the context of immune evasion.
Figures

References
-
- Adachi, O., T. Kawai, K. Takeda, M. Matsumoto, H. Tsutsui, M. Sakagami, K. Nakanishi, and S. Akira. 1998. Targeted disruption of the MyD88 gene results in loss of IL-1- and IL-18-mediated function. Immunity 9:143-150. - PubMed
-
- Balloy, V., A. Verma, S. Kuravi, M. Si-Tahar, M. Chignard, and R. Ramphal. 2007. The role of flagellin versus motility in acute lung disease caused by Pseudomonas aeruginosa. J. Infect. Dis. 196:289-296. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

