Phytophthora infestans effector AVR3a is essential for virulence and manipulates plant immunity by stabilizing host E3 ligase CMPG1
- PMID: 20457921
- PMCID: PMC2906857
- DOI: 10.1073/pnas.0914408107
Phytophthora infestans effector AVR3a is essential for virulence and manipulates plant immunity by stabilizing host E3 ligase CMPG1
Erratum in
-
Correction for Bos et al., Phytophthora infestans effector AVR3a is essential for virulence and manipulates plant immunity by stabilizing host E3 ligase CMPG1.Proc Natl Acad Sci U S A. 2020 Nov 10;117(45):28531-28533. doi: 10.1073/pnas.2019643117. Epub 2020 Oct 26. Proc Natl Acad Sci U S A. 2020. PMID: 33106432 Free PMC article. No abstract available.
Abstract
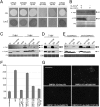
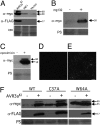
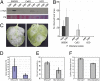
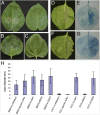
Fungal and oomycete plant pathogens translocate effector proteins into host cells to establish infection. However, virulence targets and modes of action of their effectors are unknown. Effector AVR3a from potato blight pathogen Phytophthora infestans is translocated into host cells and occurs in two forms: AVR3a(KI), which is detected by potato resistance protein R3a, strongly suppresses infestin 1 (INF1)-triggered cell death (ICD), whereas AVR3a(EM), which evades recognition by R3a, weakly suppresses host ICD. Here we show that AVR3a interacts with and stabilizes host U-box E3 ligase CMPG1, which is required for ICD. In contrast, AVR3a(KI/Y147del), a mutant with a deleted C-terminal tyrosine residue that fails to suppress ICD, cannot interact with or stabilize CMPG1. CMPG1 is stabilized by the inhibitors MG132 and epoxomicin, indicating that it is degraded by the 26S proteasome. CMPG1 is degraded during ICD. However, it is stabilized by mutations in the U-box that prevent its E3 ligase activity. In stabilizing CMPG1, AVR3a thus modifies its normal activity. Remarkably, given the potential for hundreds of effector genes in the P. infestans genome, silencing Avr3a compromises P. infestans pathogenicity, suggesting that AVR3a is essential for virulence. Interestingly, Avr3a silencing can be complemented by in planta expression of Avr3a(KI) or Avr3a(EM) but not the Avr3a(KI/Y147del) mutant. Our data provide genetic evidence that AVR3a is an essential virulence factor that targets and stabilizes the plant E3 ligase CMPG1, potentially to prevent host cell death during the biotrophic phase of infection.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Jones JD, Dangl JL. The plant immune system. Nature. 2006;444:323–329. - PubMed
-
- Chisholm ST, Coaker G, Day B, Staskawicz BJ. Host-microbe interactions: Shaping the evolution of the plant immune response. Cell. 2006;124:803–814. - PubMed
-
- Grant SR, Fisher EJ, Chang JH, Mole BM, Dangl JL. Subterfuge and manipulation: Type III effector proteins of phytopathogenic bacteria. Annu Rev Microbiol. 2006;60:425–449. - PubMed
-
- Kamoun S. A catalogue of the effector secretome of plant pathogenic oomycetes. Annu Rev Phytopathol. 2006;44:41–60. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
- BB/C006488/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/E007120/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/G015244/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BBS/B/07152/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources

