Single-base resolution mapping of H1-nucleosome interactions and 3D organization of the nucleosome
- PMID: 20457934
- PMCID: PMC2906896
- DOI: 10.1073/pnas.1000309107
Single-base resolution mapping of H1-nucleosome interactions and 3D organization of the nucleosome
Abstract
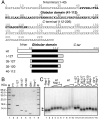
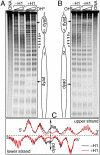
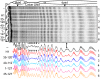
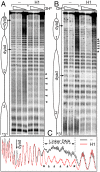
Despite the key role of the linker histone H1 in chromatin structure and dynamics, its location and interactions with nucleosomal DNA have not been elucidated. In this work we have used a combination of electron cryomicroscopy, hydroxyl radical footprinting, and nanoscale modeling to analyze the structure of precisely positioned mono-, di-, and trinucleosomes containing physiologically assembled full-length histone H1 or truncated mutants of this protein. Single-base resolution *OH footprinting shows that the globular domain of histone H1 (GH1) interacts with the DNA minor groove located at the center of the nucleosome and contacts a 10-bp region of DNA localized symmetrically with respect to the nucleosomal dyad. In addition, GH1 interacts with and organizes about one helical turn of DNA in each linker region of the nucleosome. We also find that a seven amino acid residue region (121-127) in the COOH terminus of histone H1 was required for the formation of the stem structure of the linker DNA. A molecular model on the basis of these data and coarse-grain DNA mechanics provides novel insights on how the different domains of H1 interact with the nucleosome and predicts a specific H1-mediated stem structure within linker DNA.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Structure and Dynamics of a 197 bp Nucleosome in Complex with Linker Histone H1.Mol Cell. 2017 May 4;66(3):384-397.e8. doi: 10.1016/j.molcel.2017.04.012. Mol Cell. 2017. PMID: 28475873 Free PMC article.
-
Mapping the interaction surface of linker histone H1(0) with the nucleosome of native chromatin in vivo.Nat Struct Mol Biol. 2006 Mar;13(3):250-5. doi: 10.1038/nsmb1050. Epub 2006 Feb 5. Nat Struct Mol Biol. 2006. PMID: 16462749 Free PMC article.
-
Structural insights into the histone H1-nucleosome complex.Proc Natl Acad Sci U S A. 2013 Nov 26;110(48):19390-5. doi: 10.1073/pnas.1314905110. Epub 2013 Nov 11. Proc Natl Acad Sci U S A. 2013. PMID: 24218562 Free PMC article.
-
Linker histones: novel insights into structure-specific recognition of the nucleosome.Biochem Cell Biol. 2017 Apr;95(2):171-178. doi: 10.1139/bcb-2016-0097. Epub 2016 Jun 29. Biochem Cell Biol. 2017. PMID: 28177778 Free PMC article. Review.
-
H1-nucleosome interactions and their functional implications.Biochim Biophys Acta. 2016 Mar;1859(3):436-43. doi: 10.1016/j.bbagrm.2015.10.012. Epub 2015 Oct 23. Biochim Biophys Acta. 2016. PMID: 26477489 Review.
Cited by
-
Genome-wide characterization and evolutionary analysis of linker histones in castor bean (Ricinus communis).Front Plant Sci. 2022 Oct 21;13:1014418. doi: 10.3389/fpls.2022.1014418. eCollection 2022. Front Plant Sci. 2022. PMID: 36340363 Free PMC article.
-
Structural basis of a nucleosome containing histone H2A.B/H2A.Bbd that transiently associates with reorganized chromatin.Sci Rep. 2013 Dec 16;3:3510. doi: 10.1038/srep03510. Sci Rep. 2013. PMID: 24336483 Free PMC article.
-
Base excision repair of 8-oxoG in dinucleosomes.Nucleic Acids Res. 2012 Jan;40(2):692-700. doi: 10.1093/nar/gkr761. Epub 2011 Sep 19. Nucleic Acids Res. 2012. PMID: 21930508 Free PMC article.
-
Dependence of Chromatosome Structure on Linker Histone Sequence and Posttranslational Modification.Biophys J. 2018 May 22;114(10):2363-2375. doi: 10.1016/j.bpj.2018.04.034. Epub 2018 May 11. Biophys J. 2018. PMID: 29759374 Free PMC article.
-
Elucidating the influence of linker histone variants on chromatosome dynamics and energetics.Nucleic Acids Res. 2020 Apr 17;48(7):3591-3604. doi: 10.1093/nar/gkaa121. Nucleic Acids Res. 2020. PMID: 32128577 Free PMC article.
References
-
- Luger K, Mäder AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. - PubMed
-
- Hamiche A, Schultz P, Ramakrishnan V, Oudet P, Prunell A. Linker histone-dependent DNA structure in linear mononucleosomes. J Mol Biol. 1996;257(1):30–42. - PubMed
-
- Russanova VR, Dimitrov SI, Makarov VL, Pashev IG. Accessibility of the globular domain of histones H1 and H5 to antibodies upon folding of chromatin. Eur J Biochem. 1987;167(2):321–326. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

