Inference of cancer-specific gene regulatory networks using soft computing rules
- PMID: 20458373
- PMCID: PMC2865768
- DOI: 10.4137/grsb.s4509
Inference of cancer-specific gene regulatory networks using soft computing rules
Abstract
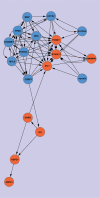
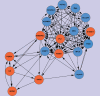
Perturbations of gene regulatory networks are essentially responsible for oncogenesis. Therefore, inferring the gene regulatory networks is a key step to overcoming cancer. In this work, we propose a method for inferring directed gene regulatory networks based on soft computing rules, which can identify important cause-effect regulatory relations of gene expression. First, we identify important genes associated with a specific cancer (colon cancer) using a supervised learning approach. Next, we reconstruct the gene regulatory networks by inferring the regulatory relations among the identified genes, and their regulated relations by other genes within the genome. We obtain two meaningful findings. One is that upregulated genes are regulated by more genes than downregulated ones, while downregulated genes regulate more genes than upregulated ones. The other one is that tumor suppressors suppress tumor activators and activate other tumor suppressors strongly, while tumor activators activate other tumor activators and suppress tumor suppressors weakly, indicating the robustness of biological systems. These findings provide valuable insights into the pathogenesis of cancer.
Keywords: cancer; decision rules; gene regulatory networks; machine learning; microarrays.
Figures

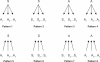


References
-
- Kitano H. Systems biology: a brief overview. Science. 2002;295(5560):1662–4. - PubMed
-
- Hornberg JJ, Bruggeman FJ, Westerhoff HV, Lankelma J. Cancer: a systems biology disease. Biosystems. 2006;83:2–3. 81–90. - PubMed
-
- Vogelstein B, Kinzler KW. Cancer genes and the pathways they control. Nat Med. 2004;10(8):789–99. - PubMed
-
- Awan A, Bari H, Yan F, et al. Regulatory network motifs and hotspots of cancer genes in a mammalian cellular signalling network. IET Syst Biol. 2007;1(5):292–7. - PubMed
LinkOut - more resources
Full Text Sources

