Non-canonical activation of Notch signaling/target genes in vertebrates
- PMID: 20458516
- PMCID: PMC11115867
- DOI: 10.1007/s00018-010-0391-x
Non-canonical activation of Notch signaling/target genes in vertebrates
Abstract
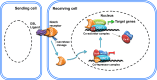
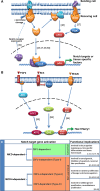
Evolutionarily conserved Notch signaling orchestrates diverse physiological mechanisms during metazoan development and homeostasis. Classically, ligand-activated Notch receptors transduce the signaling cascade through the interaction of DNA-bound CBF1-co-repressor complex. However, recent reports have demonstrated execution of a CBF1-independent Notch pathway through signaling cross-talks in various cells/tissues. Here, we have tried to congregate the reports that describe the non-canonical/CBF1-independent Notch signaling and target gene activation in vertebrates with specific emphasis on their functional relevance.
Figures
References
-
- Artavanis-Tsakonas S, Rand MD, Lake RJ. Notch signaling: cell fate control and signal integration in development. Science. 1999;284:770–776. - PubMed
-
- Mumm JS, Kopan R. Notch signaling: from the outside in. Dev Biol. 2000;228:151–165. - PubMed
-
- Honjo T. The shortest path from the surface to the nucleus: RBP-J kappa/Su(H) transcription factor. Genes Cells. 1996;1:1–9. - PubMed
-
- Bray SJ. Notch signalling: a simple pathway becomes complex. Nat Rev Mol Cell Biol. 2006;7:678–689. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

