Promoter specificity and interactions between early and late Arabidopsis heat shock factors
- PMID: 20458611
- PMCID: PMC2882041
- DOI: 10.1007/s11103-010-9643-2
Promoter specificity and interactions between early and late Arabidopsis heat shock factors
Abstract
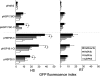
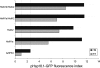
The class A heat shock factors HsfA1a and HsfA1b are highly conserved, interacting regulators, responsible for the immediate-early transcription of a subset of heat shock genes in Arabidopsis. In order to determine functional cooperation between them, we used a reporter assay based on transient over-expression in Arabidopsis protoplasts. Reporter plasmids containing promoters of Hsf target genes fused with the GFP coding region were co-transformed with Hsf effector plasmids. The GFP reporter gene activity was quantified using flow cytometry. Three of the tested target gene promoters (Hsp25.3, Hsp18.1-CI, Hsp26.5) resulted in a strong reporter gene activity, with HsfA1a or HsfA1b alone, and significantly enhanced GFP fluorescence when both effectors were co-transformed. A second set of heat shock promoters (HsfA2, Hsp17.6CII, Hsp17.6C-CI) was activated to much lower levels. These data suggest that HsfA1a/1b cooperate synergistically at a number of target gene promoters. These targets are also regulated via the late HsfA2, which is the most strongly heat-induced class A-Hsf in Arabidopsis. HsfA2 has also the capacity to interact with HsfA1a and HsfA1b as determined by bimolecular fluorescence complementation (BiFC) in Arabidopsis protoplasts and yeast-two-hybrid assay. However, there was no synergistic effect on Hsp18.1-CI promoter-GFP reporter gene expression when HsfA2 was co-expressed with either HsfA1a or HsfA1b. These data provide evidence that interaction between early and late HSF is possible, but only interaction between the early Hsfs results in a synergistic enhancement of expression of certain target genes. The interaction of HsfA1a/A1b with the major-late HsfA2 may possibly support recruitment of HsfA2 and replacement of HsfA1a/A1b at the same target gene promoters.
Figures


Similar articles
-
Detection of in vivo interactions between Arabidopsis class A-HSFs, using a novel BiFC fragment, and identification of novel class B-HSF interacting proteins.Eur J Cell Biol. 2010 Feb-Mar;89(2-3):126-32. doi: 10.1016/j.ejcb.2009.10.012. Epub 2009 Nov 27. Eur J Cell Biol. 2010. PMID: 19945192
-
Arabidopsis heat shock factor HsfA1a directly senses heat stress, pH changes, and hydrogen peroxide via the engagement of redox state.Plant Physiol Biochem. 2013 Mar;64:92-8. doi: 10.1016/j.plaphy.2012.12.013. Epub 2013 Jan 5. Plant Physiol Biochem. 2013. PMID: 23399534
-
The heat-inducible transcription factor HsfA2 enhances anoxia tolerance in Arabidopsis.Plant Physiol. 2010 Mar;152(3):1471-83. doi: 10.1104/pp.109.149815. Epub 2010 Jan 20. Plant Physiol. 2010. PMID: 20089772 Free PMC article.
-
Heat stress response in plants: a complex game with chaperones and more than twenty heat stress transcription factors.J Biosci. 2004 Dec;29(4):471-87. doi: 10.1007/BF02712120. J Biosci. 2004. PMID: 15625403 Review.
-
Arabidopsis and the heat stress transcription factor world: how many heat stress transcription factors do we need?Cell Stress Chaperones. 2001 Jul;6(3):177-89. doi: 10.1379/1466-1268(2001)006<0177:aathst>2.0.co;2. Cell Stress Chaperones. 2001. PMID: 11599559 Free PMC article. Review.
Cited by
-
Overall picture of expressed Heat Shock Factors in Glycine max, Lotus japonicus and Medicago truncatula.Genet Mol Biol. 2012 Jun;35(1 (suppl)):247-59. doi: 10.1590/S1415-47572012000200006. Genet Mol Biol. 2012. PMID: 22802710 Free PMC article.
-
Heat reduces nitric oxide production required for auxin-mediated gene expression and fate determination in tree tobacco guard cell protoplasts.Plant Physiol. 2012 Aug;159(4):1608-23. doi: 10.1104/pp.112.200089. Epub 2012 Jun 22. Plant Physiol. 2012. PMID: 22730424 Free PMC article.
-
Priming by High Temperature Stress Induces MicroRNA Regulated Heat Shock Modules Indicating Their Involvement in Thermopriming Response in Rice.Life (Basel). 2021 Mar 29;11(4):291. doi: 10.3390/life11040291. Life (Basel). 2021. PMID: 33805566 Free PMC article.
-
Arabidopsis HEAT SHOCK TRANSCRIPTION FACTORA1b overexpression enhances water productivity, resistance to drought, and infection.J Exp Bot. 2013 Aug;64(11):3467-81. doi: 10.1093/jxb/ert185. Epub 2013 Jul 4. J Exp Bot. 2013. PMID: 23828547 Free PMC article.
-
Characterization and Functional Analysis of FaHsfC1b from Festuca arundinacea Conferring Heat Tolerance in Arabidopsis.Int J Mol Sci. 2018 Sep 11;19(9):2702. doi: 10.3390/ijms19092702. Int J Mol Sci. 2018. PMID: 30208588 Free PMC article.
References
-
- Bharti K, von Koskull-Döring P, Bharti S, Kumar P, Tintschl-Korbitzer A, Treuter E, Nover L. Tomato heat stress transcrition factor HsfB1 represents a novel type of general transcription coactivator with a histone-like motif interacting with the plant CREB binding protein ortholog HAC1. Plant Cell. 2004;16:1521–1535. doi: 10.1105/tpc.019927. - DOI - PMC - PubMed
-
- Chan-Schaminet KY, Baniwal SK, Bublak D, Nover L, Scharf K-D. Specific interaction between tomato HsfA1 and HsfA2 creates hetero-oligomeric superactivator complexes for synergistic activation of heat stress gene expression. J Biol Chem. 2009;284:20848–20857. doi: 10.1074/jbc.M109.007336. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

