Copy number variation in the bovine genome
- PMID: 20459598
- PMCID: PMC2902221
- DOI: 10.1186/1471-2164-11-284
Copy number variation in the bovine genome
Abstract
Background: Copy number variations (CNVs), which represent a significant source of genetic diversity in mammals, have been shown to be associated with phenotypes of clinical relevance and to be causative of disease. Notwithstanding, little is known about the extent to which CNV contributes to genetic variation in cattle.
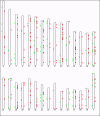
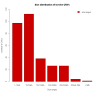
Results: We designed and used a set of NimbleGen CGH arrays that tile across the assayable portion of the cattle genome with approximately 6.3 million probes, at a median probe spacing of 301 bp. This study reports the highest resolution map of copy number variation in the cattle genome, with 304 CNV regions (CNVRs) being identified among the genomes of 20 bovine samples from 4 dairy and beef breeds. The CNVRs identified covered 0.68% (22 Mb) of the genome, and ranged in size from 1.7 to 2,031 kb (median size 16.7 kb). About 20% of the CNVs co-localized with segmental duplications, while 30% encompass genes, of which the majority is involved in environmental response. About 10% of the human orthologous of these genes are associated with human disease susceptibility and, hence, may have important phenotypic consequences.
Conclusions: Together, this analysis provides a useful resource for assessment of the impact of CNVs regarding variation in bovine health and production traits.
Figures


Similar articles
-
An initial comparative map of copy number variations in the goat (Capra hircus) genome.BMC Genomics. 2010 Nov 17;11:639. doi: 10.1186/1471-2164-11-639. BMC Genomics. 2010. PMID: 21083884 Free PMC article.
-
Identification of copy number variations in Qinchuan cattle using BovineHD Genotyping Beadchip array.Mol Genet Genomics. 2015 Feb;290(1):319-27. doi: 10.1007/s00438-014-0923-4. Epub 2014 Sep 24. Mol Genet Genomics. 2015. PMID: 25248638
-
Analysis of copy number variations among diverse cattle breeds.Genome Res. 2010 May;20(5):693-703. doi: 10.1101/gr.105403.110. Epub 2010 Mar 8. Genome Res. 2010. PMID: 20212021 Free PMC article.
-
Copy number variation in the cattle genome.Funct Integr Genomics. 2012 Nov;12(4):609-24. doi: 10.1007/s10142-012-0289-9. Epub 2012 Jul 13. Funct Integr Genomics. 2012. PMID: 22790923 Review.
-
Copy number variation in the domestic dog.Mamm Genome. 2012 Feb;23(1-2):144-63. doi: 10.1007/s00335-011-9369-8. Epub 2011 Dec 4. Mamm Genome. 2012. PMID: 22138850 Review.
Cited by
-
Genome-wide identification of copy number variations in Chinese Holstein.PLoS One. 2012;7(11):e48732. doi: 10.1371/journal.pone.0048732. Epub 2012 Nov 7. PLoS One. 2012. PMID: 23144949 Free PMC article.
-
Diversity and population-genetic properties of copy number variations and multicopy genes in cattle.DNA Res. 2016 Jun;23(3):253-62. doi: 10.1093/dnares/dsw013. Epub 2016 Apr 15. DNA Res. 2016. PMID: 27085184 Free PMC article.
-
Genome-wide identification of copy number variation and association with fat deposition in thin and fat-tailed sheep breeds.Sci Rep. 2022 May 25;12(1):8834. doi: 10.1038/s41598-022-12778-1. Sci Rep. 2022. PMID: 35614300 Free PMC article.
-
Detection of Copy Number Variations in Woori-Heukdon Populations with the Illumina PorcineSNP60 Bead-Chip Array.Animals (Basel). 2025 Mar 9;15(6):774. doi: 10.3390/ani15060774. Animals (Basel). 2025. PMID: 40150303 Free PMC article.
-
Testis-Specific Protein Y-Encoded (TSPY) Is Required for Male Early Embryo Development in Bos taurus.Int J Mol Sci. 2023 Feb 8;24(4):3349. doi: 10.3390/ijms24043349. Int J Mol Sci. 2023. PMID: 36834761 Free PMC article.
References
-
- Lejeune J, Gautier M, Turpin R. Study of somatic chromosomes from 9 mongoloid children. C R Hebd Seances Acad Sci. 1959;248:1721–1722. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

