MUC1-associated proliferation signature predicts outcomes in lung adenocarcinoma patients
- PMID: 20459602
- PMCID: PMC2876055
- DOI: 10.1186/1755-8794-3-16
MUC1-associated proliferation signature predicts outcomes in lung adenocarcinoma patients
Abstract
Background: MUC1 protein is highly expressed in lung cancer. The cytoplasmic domain of MUC1 (MUC1-CD) induces tumorigenesis and resistance to DNA-damaging agents. We characterized MUC1-CD-induced transcriptional changes and examined their significance in lung cancer patients.
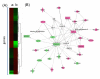
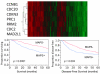
Methods: Using DNA microarrays, we identified 254 genes that were differentially expressed in cell lines transformed by MUC1-CD compared to control cell lines. We then examined expression of these genes in 441 lung adenocarcinomas from a publicly available database. We employed statistical analyses independent of clinical outcomes, including hierarchical clustering, Student's t-tests and receiver operating characteristic (ROC) analysis, to select a seven-gene MUC1-associated proliferation signature (MAPS). We demonstrated the prognostic value of MAPS in this database using Kaplan-Meier survival analysis, log-rank tests and Cox models. The MAPS was further validated for prognostic significance in 84 lung adenocarcinoma patients from an independent database.
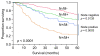
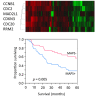
Results: MAPS genes were found to be associated with proliferation and cell cycle regulation and included CCNB1, CDC2, CDC20, CDKN3, MAD2L1, PRC1 and RRM2. MAPS expressors (MAPS+) had inferior survival compared to non-expressors (MAPS-). In the initial data set, 5-year survival was 65% (MAPS-) vs. 45% (MAPS+, p < 0.0001). Similarly, in the validation data set, 5-year survival was 57% (MAPS-) vs. 28% (MAPS+, p = 0.005).
Conclusions: The MAPS signature, comprised of MUC1-CD-dependent genes involved in the control of cell cycle and proliferation, is associated with poor outcomes in patients with adenocarcinoma of the lung. These data provide potential new prognostic biomarkers and treatment targets for lung adenocarcinoma.
Figures


Similar articles
-
Targeting the intracellular MUC1 C-terminal domain inhibits proliferation and estrogen receptor transcriptional activity in lung adenocarcinoma cells.Mol Cancer Ther. 2011 Nov;10(11):2062-71. doi: 10.1158/1535-7163.MCT-11-0381. Epub 2011 Aug 23. Mol Cancer Ther. 2011. PMID: 21862684 Free PMC article.
-
Mucin-1 correlates with survival, smoking status, and growth patterns in lung adenocarcinoma.Tumour Biol. 2016 Oct;37(10):13811-13820. doi: 10.1007/s13277-016-5269-6. Epub 2016 Aug 1. Tumour Biol. 2016. PMID: 27481516
-
MUC1-induced transcriptional programs associated with tumorigenesis predict outcome in breast and lung cancer.Cancer Res. 2009 Apr 1;69(7):2833-7. doi: 10.1158/0008-5472.CAN-08-4513. Epub 2009 Mar 24. Cancer Res. 2009. PMID: 19318547 Free PMC article.
-
Expression and prognostic relevance of MUC1 in stage IB non-small cell lung cancer.Med Oncol. 2011 Dec;28 Suppl 1:S596-604. doi: 10.1007/s12032-010-9752-4. Epub 2010 Nov 30. Med Oncol. 2011. PMID: 21116877
-
Development and validation of a robust immune-related prognostic signature in early-stage lung adenocarcinoma.J Transl Med. 2020 Oct 7;18(1):380. doi: 10.1186/s12967-020-02545-z. J Transl Med. 2020. PMID: 33028329 Free PMC article.
Cited by
-
Mucin Expression in Mucinous Pancreatic Cysts: Can String Sign Test Predict Mucin Types? A Single Center Pilot Study.Turk J Gastroenterol. 2021 Feb;32(2):187-193. doi: 10.5152/tjg.2020.20060. Turk J Gastroenterol. 2021. PMID: 33960943 Free PMC article.
-
An algorithm to quantify intratumor heterogeneity based on alterations of gene expression profiles.Commun Biol. 2020 Sep 11;3(1):505. doi: 10.1038/s42003-020-01230-7. Commun Biol. 2020. PMID: 32917965 Free PMC article.
-
Smoking-induced CXCL14 expression in the human airway epithelium links chronic obstructive pulmonary disease to lung cancer.Am J Respir Cell Mol Biol. 2013 Sep;49(3):418-25. doi: 10.1165/rcmb.2012-0396OC. Am J Respir Cell Mol Biol. 2013. PMID: 23597004 Free PMC article.
-
TRPV1 Is a Potential Tumor Suppressor for Its Negative Association with Tumor Proliferation and Positive Association with Antitumor Immune Responses in Pan-Cancer.J Oncol. 2022 Oct 18;2022:6964550. doi: 10.1155/2022/6964550. eCollection 2022. J Oncol. 2022. PMID: 36304985 Free PMC article.
-
MUC1-C Is a Common Driver of Acquired Osimertinib Resistance in NSCLC.J Thorac Oncol. 2024 Mar;19(3):434-450. doi: 10.1016/j.jtho.2023.10.017. Epub 2023 Nov 3. J Thorac Oncol. 2024. PMID: 37924972 Free PMC article.
References
-
- Govindan R, Page N, Morgensztern D, Read W, Tierney R, Vlahiotis A, Spitznagel EL, Piccirillo J. Changing epidemiology of small-cell lung cancer in the United States over the last 30 years: analysis of the surveillance, epidemiologic, and end results database. J Clin Oncol. 2006;24(28):4539–4544. doi: 10.1200/JCO.2005.04.4859. - DOI - PubMed
-
- Auperin A, Le Pechoux C, Pignon JP, Koning C, Jeremic B, Clamon G, Einhorn L, Ball D, Trovo MG, Groen HJ. Concomitant radio-chemotherapy based on platin compounds in patients with locally advanced non-small cell lung cancer (NSCLC): a meta-analysis of individual data from 1764 patients. Ann Oncol. 2006;17(3):473–483. doi: 10.1093/annonc/mdj117. - DOI - PubMed
-
- Tsao MS, Zhu C, Ding K, Strumpf D, Pintilie M, Meyerson M, Seymour L, Jurisica I, Shepherd FA. A 15-gene expression signature prognostic for survival and predictive for adjuvant chemotherapy benefit in JBR.10 patients. J Clin Oncol. 2008;26 (May 20 suppl; abstr 7510)
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

