High-resolution genomic and expression analyses of copy number alterations in HER2-amplified breast cancer
- PMID: 20459607
- PMCID: PMC2917012
- DOI: 10.1186/bcr2568
High-resolution genomic and expression analyses of copy number alterations in HER2-amplified breast cancer
Abstract
Introduction: HER2 gene amplification and protein overexpression (HER2+) define a clinically challenging subgroup of breast cancer with variable prognosis and response to therapy. Although gene expression profiling has identified an ERBB2 molecular subtype of breast cancer, it is clear that HER2+ tumors reside in all molecular subtypes and represent a genomically and biologically heterogeneous group, needed to be further characterized in large sample sets.
Methods: Genome-wide DNA copy number profiling, using bacterial artificial chromosome (BAC) array comparative genomic hybridization (aCGH), and global gene expression profiling were performed on 200 and 87 HER2+ tumors, respectively. Genomic Identification of Significant Targets in Cancer (GISTIC) was used to identify significant copy number alterations (CNAs) in HER2+ tumors, which were related to a set of 554 non-HER2 amplified (HER2-) breast tumors. High-resolution oligonucleotide aCGH was used to delineate the 17q12-q21 region in high detail.
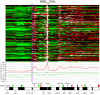
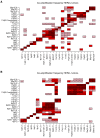
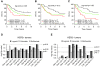
Results: The HER2-amplicon was narrowed to an 85.92 kbp region including the TCAP, PNMT, PERLD1, HER2, C17orf37 and GRB7 genes, and higher HER2 copy numbers indicated worse prognosis. In 31% of HER2+ tumors the amplicon extended to TOP2A, defining a subgroup of HER2+ breast cancer associated with estrogen receptor-positive status and with a trend of better survival than HER2+ breast cancers with deleted (18%) or neutral TOP2A (51%). HER2+ tumors were clearly distinguished from HER2- tumors by the presence of recurrent high-level amplifications and firestorm patterns on chromosome 17q. While there was no significant difference between HER2+ and HER2- tumors regarding the incidence of other recurrent high-level amplifications, differences in the co-amplification pattern were observed, as shown by the almost mutually exclusive occurrence of 8p12, 11q13 and 20q13 amplification in HER2+ tumors. GISTIC analysis identified 117 significant CNAs across all autosomes. Supervised analyses revealed: (1) significant CNAs separating HER2+ tumors stratified by clinical variables, and (2) CNAs separating HER2+ from HER2- tumors.
Conclusions: We have performed a comprehensive survey of CNAs in HER2+ breast tumors, pinpointing significant genomic alterations including both known and potentially novel therapeutic targets. Our analysis sheds further light on the genomically complex and heterogeneous nature of HER2+ tumors in relation to other subgroups of breast cancer.
Figures


Comment in
-
What do we learn from HER2-positive breast cancer genomic profiles?Breast Cancer Res. 2010;12(3):107. doi: 10.1186/bcr2571. Epub 2010 Jun 1. Breast Cancer Res. 2010. PMID: 20519027 Free PMC article.
References
-
- Jonkers J, Berns A. Oncogene addiction: sometimes a temporary slavery. Cancer Cell. 2004;6:535–538. - PubMed
-
- Paik S, Hazan R, Fisher ER, Sass RE, Fisher B, Redmond C, Schlessinger J, Lippman ME, King CR. Pathologic findings from the National Surgical Adjuvant Breast and Bowel Project: prognostic significance of erbB-2 protein overexpression in primary breast cancer. J Clin Oncol. 1990;8:103–112. - PubMed
-
- Smith I, Procter M, Gelber RD, Guillaume S, Feyereislova A, Dowsett M, Goldhirsch A, Untch M, Mariani G, Baselga J, Kaufmann M, Cameron D, Bell R, Bergh J, Coleman R, Wardley A, Harbeck N, Lopez RI, Mallmann P, Gelmon K, Wilcken N, Wist E, Sanchez Rovira P, Piccart-Gebhart MJ. 2-year follow-up of trastuzumab after adjuvant chemotherapy in HER2-positive breast cancer: a randomised controlled trial. Lancet. 2007;369:29–36. doi: 10.1016/S0140-6736(07)60028-2. - DOI - PubMed
-
- Romond EH, Perez EA, Bryant J, Suman VJ, Geyer CE Jr, Davidson NE, Tan-Chiu E, Martino S, Paik S, Kaufman PA, Swain SM, Pisansky TM, Fehrenbacher L, Kutteh LA, Vogel VG, Visscher DW, Yothers G, Jenkins RB, Brown AM, Dakhil SR, Mamounas EP, Lingle WL, Klein PM, Ingle JN, Wolmark N. Trastuzumab plus adjuvant chemotherapy for operable HER2-positive breast cancer. N Engl J Med. 2005;353:1673–1684. doi: 10.1056/NEJMoa052122. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

