Frequent and recent retrotransposition of orthologous genes plays a role in the evolution of sperm glycolytic enzymes
- PMID: 20459611
- PMCID: PMC2881024
- DOI: 10.1186/1471-2164-11-285
Frequent and recent retrotransposition of orthologous genes plays a role in the evolution of sperm glycolytic enzymes
Abstract
Background: The central metabolic pathway of glycolysis converts glucose to pyruvate, with the net production of 2 ATP and 2 NADH per glucose molecule. Each of the ten reactions in this pathway is typically catalyzed by multiple isozymes encoded by a multigene family. Several isozymes in this pathway are expressed only during spermatogenesis, and gene targeting studies indicate that they are essential for sperm function and male fertility in mouse. At least three of the novel glycolytic isozymes are encoded by retrogenes (Pgk2, Aldoart1, and Aldoart2). Their restricted expression profile suggests that retrotransposition may play a significant role in the evolution of sperm glycolytic enzymes.
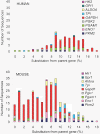
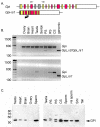
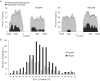
Results: We conducted a comprehensive genomic analysis of glycolytic enzymes in the human and mouse genomes and identified several intronless copies for all enzymes in the pathway, except Pfk. Within each gene family, a single orthologous gene was typically retrotransposed frequently and independently in both species. Several retroposed sequences maintained open reading frames (ORFs) and/or provided evidence of alternatively spliced exons. We analyzed expression of sequences with ORFs and <99% sequence identity in the coding region and obtained evidence for the expression of an alternative Gpi1 transcript in mouse spermatogenic cells.
Conclusions: Our analysis detected frequent, recent, and lineage-specific retrotransposition of orthologous glycolytic enzymes in the human and mouse genomes. Retrotransposition events are associated with LINE/LTR and genomic integration is random. We found evidence for the alternative splicing of parent genes. Many retroposed sequences have maintained ORFs, suggesting a functional role for these genes.
Figures


References
-
- Mori C, Nakamura N, Welch JE, Gotoh H, Goulding EH, Fujioka M, Eddy EM. Mouse spermatogenic cell-specific type 1 hexokinase (mHk1-s) transcripts are expressed by alternative splicing from the mHk1 gene and the HK1-S protein is localized mainly in the sperm tail. Mol Reprod Dev. 1998;49:374–385. doi: 10.1002/(SICI)1098-2795(199804)49:4<374::AID-MRD4>3.0.CO;2-K. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

