RNA FRABASE 2.0: an advanced web-accessible database with the capacity to search the three-dimensional fragments within RNA structures
- PMID: 20459631
- PMCID: PMC2873543
- DOI: 10.1186/1471-2105-11-231
RNA FRABASE 2.0: an advanced web-accessible database with the capacity to search the three-dimensional fragments within RNA structures
Abstract
Background: Recent discoveries concerning novel functions of RNA, such as RNA interference, have contributed towards the growing importance of the field. In this respect, a deeper knowledge of complex three-dimensional RNA structures is essential to understand their new biological functions. A number of bioinformatic tools have been proposed to explore two major structural databases (PDB, NDB) in order to analyze various aspects of RNA tertiary structures. One of these tools is RNA FRABASE 1.0, the first web-accessible database with an engine for automatic search of 3D fragments within PDB-derived RNA structures. This search is based upon the user-defined RNA secondary structure pattern. In this paper, we present and discuss RNA FRABASE 2.0. This second version of the system represents a major extension of this tool in terms of providing new data and a wide spectrum of novel functionalities. An intuitionally operated web server platform enables very fast user-tailored search of three-dimensional RNA fragments, their multi-parameter conformational analysis and visualization.
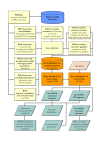
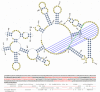
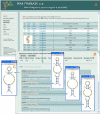
Description: RNA FRABASE 2.0 has stored information on 1565 PDB-deposited RNA structures, including all NMR models. The RNA FRABASE 2.0 search engine algorithms operate on the database of the RNA sequences and the new library of RNA secondary structures, coded in the dot-bracket format extended to hold multi-stranded structures and to cover residues whose coordinates are missing in the PDB files. The library of RNA secondary structures (and their graphics) is made available. A high level of efficiency of the 3D search has been achieved by introducing novel tools to formulate advanced searching patterns and to screen highly populated tertiary structure elements. RNA FRABASE 2.0 also stores data and conformational parameters in order to provide "on the spot" structural filters to explore the three-dimensional RNA structures. An instant visualization of the 3D RNA structures is provided. RNA FRABASE 2.0 is freely available at http://rnafrabase.cs.put.poznan.pl.
Conclusions: RNA FRABASE 2.0 provides a novel database and powerful search engine which is equipped with new data and functionalities that are unavailable elsewhere. Our intention is that this advanced version of the RNA FRABASE will be of interest to all researchers working in the RNA field.
Figures



Similar articles
-
RNA FRABASE version 1.0: an engine with a database to search for the three-dimensional fragments within RNA structures.Nucleic Acids Res. 2008 Jan;36(Database issue):D386-91. doi: 10.1093/nar/gkm786. Epub 2007 Oct 5. Nucleic Acids Res. 2008. PMID: 17921499 Free PMC article.
-
R3D-BLAST2: an improved search tool for similar RNA 3D substructures.BMC Bioinformatics. 2017 Dec 28;18(Suppl 16):574. doi: 10.1186/s12859-017-1956-6. BMC Bioinformatics. 2017. PMID: 29297283 Free PMC article.
-
Automated 3D structure composition for large RNAs.Nucleic Acids Res. 2012 Aug;40(14):e112. doi: 10.1093/nar/gks339. Epub 2012 Apr 26. Nucleic Acids Res. 2012. PMID: 22539264 Free PMC article.
-
Design and simulation of DNA, RNA and hybrid protein-nucleic acid nanostructures with oxView.Nat Protoc. 2022 Aug;17(8):1762-1788. doi: 10.1038/s41596-022-00688-5. Epub 2022 Jun 6. Nat Protoc. 2022. PMID: 35668321 Review.
-
Software tools for visualizing Hi-C data.Genome Biol. 2017 Feb 3;18(1):26. doi: 10.1186/s13059-017-1161-y. Genome Biol. 2017. PMID: 28159004 Free PMC article. Review.
Cited by
-
Nucleic acid helix structure determination from NMR proton chemical shifts.J Biomol NMR. 2013 Jun;56(2):95-112. doi: 10.1007/s10858-013-9725-y. Epub 2013 Apr 6. J Biomol NMR. 2013. PMID: 23564038
-
NASSAM: a server to search for and annotate tertiary interactions and motifs in three-dimensional structures of complex RNA molecules.Nucleic Acids Res. 2012 Jul;40(Web Server issue):W35-41. doi: 10.1093/nar/gks513. Epub 2012 May 31. Nucleic Acids Res. 2012. PMID: 22661578 Free PMC article.
-
RAG-3D: a search tool for RNA 3D substructures.Nucleic Acids Res. 2015 Oct 30;43(19):9474-88. doi: 10.1093/nar/gkv823. Epub 2015 Aug 24. Nucleic Acids Res. 2015. PMID: 26304547 Free PMC article.
-
Computational Methods for Modeling Aptamers and Designing Riboswitches.Int J Mol Sci. 2017 Nov 17;18(11):2442. doi: 10.3390/ijms18112442. Int J Mol Sci. 2017. PMID: 29149090 Free PMC article. Review.
-
AmiR-P3: An AI-based microRNA prediction pipeline in plants.PLoS One. 2024 Aug 1;19(8):e0308016. doi: 10.1371/journal.pone.0308016. eCollection 2024. PLoS One. 2024. PMID: 39088479 Free PMC article.
References
-
- Berman HM, Westbrook J, Feng Z, Iype L, Schneider B, Zardecki C. The nucleic acid database. Methods Biochem Anal. 2003;44:199–216. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

