Odefy--from discrete to continuous models
- PMID: 20459647
- PMCID: PMC2873544
- DOI: 10.1186/1471-2105-11-233
Odefy--from discrete to continuous models
Abstract
Background: Phenomenological information about regulatory interactions is frequently available and can be readily converted to Boolean models. Fully quantitative models, on the other hand, provide detailed insights into the precise dynamics of the underlying system. In order to connect discrete and continuous modeling approaches, methods for the conversion of Boolean systems into systems of ordinary differential equations have been developed recently. As biological interaction networks have steadily grown in size and complexity, a fully automated framework for the conversion process is desirable.
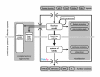
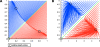
Results: We present Odefy, a MATLAB- and Octave-compatible toolbox for the automated transformation of Boolean models into systems of ordinary differential equations. Models can be created from sets of Boolean equations or graph representations of Boolean networks. Alternatively, the user can import Boolean models from the CellNetAnalyzer toolbox, GINSim and the PBN toolbox. The Boolean models are transformed to systems of ordinary differential equations by multivariate polynomial interpolation and optional application of sigmoidal Hill functions. Our toolbox contains basic simulation and visualization functionalities for both, the Boolean as well as the continuous models. For further analyses, models can be exported to SQUAD, GNA, MATLAB script files, the SB toolbox, SBML and R script files. Odefy contains a user-friendly graphical user interface for convenient access to the simulation and exporting functionalities. We illustrate the validity of our transformation approach as well as the usage and benefit of the Odefy toolbox for two biological systems: a mutual inhibitory switch known from stem cell differentiation and a regulatory network giving rise to a specific spatial expression pattern at the mid-hindbrain boundary.
Conclusions: Odefy provides an easy-to-use toolbox for the automatic conversion of Boolean models to systems of ordinary differential equations. It can be efficiently connected to a variety of input and output formats for further analysis and investigations. The toolbox is open-source and can be downloaded at http://cmb.helmholtz-muenchen.de/odefy.
Figures



Similar articles
-
Boolean network modeling in systems pharmacology.J Pharmacokinet Pharmacodyn. 2018 Feb;45(1):159-180. doi: 10.1007/s10928-017-9567-4. Epub 2018 Jan 6. J Pharmacokinet Pharmacodyn. 2018. PMID: 29307099 Free PMC article. Review.
-
CRA toolbox: software package for conditional robustness analysis of cancer systems biology models in MATLAB.BMC Bioinformatics. 2019 Jul 9;20(1):385. doi: 10.1186/s12859-019-2933-z. BMC Bioinformatics. 2019. PMID: 31288758 Free PMC article.
-
optPBN: an optimisation toolbox for probabilistic Boolean networks.PLoS One. 2014 Jul 1;9(7):e98001. doi: 10.1371/journal.pone.0098001. eCollection 2014. PLoS One. 2014. PMID: 24983623 Free PMC article.
-
ASSA-PBN: A Toolbox for Probabilistic Boolean Networks.IEEE/ACM Trans Comput Biol Bioinform. 2018 Jul-Aug;15(4):1203-1216. doi: 10.1109/TCBB.2017.2773477. Epub 2017 Nov 14. IEEE/ACM Trans Comput Biol Bioinform. 2018. PMID: 29990128
-
Boolean modeling in systems biology: an overview of methodology and applications.Phys Biol. 2012 Oct;9(5):055001. doi: 10.1088/1478-3975/9/5/055001. Epub 2012 Sep 25. Phys Biol. 2012. PMID: 23011283 Review.
Cited by
-
A Network-Based Data Integration Approach to Support Drug Repurposing and Multi-Target Therapies in Triple Negative Breast Cancer.PLoS One. 2016 Sep 15;11(9):e0162407. doi: 10.1371/journal.pone.0162407. eCollection 2016. PLoS One. 2016. PMID: 27632168 Free PMC article.
-
Boolean network modeling in systems pharmacology.J Pharmacokinet Pharmacodyn. 2018 Feb;45(1):159-180. doi: 10.1007/s10928-017-9567-4. Epub 2018 Jan 6. J Pharmacokinet Pharmacodyn. 2018. PMID: 29307099 Free PMC article. Review.
-
A parallel attractor-finding algorithm based on Boolean satisfiability for genetic regulatory networks.PLoS One. 2014 Apr 9;9(4):e94258. doi: 10.1371/journal.pone.0094258. eCollection 2014. PLoS One. 2014. PMID: 24718686 Free PMC article.
-
Convergence behaviour and Control in Non-Linear Biological Networks.Sci Rep. 2015 Jun 11;5:9746. doi: 10.1038/srep09746. Sci Rep. 2015. PMID: 26068060 Free PMC article.
-
Simulation of the Dynamics of Primary Immunodeficiencies in B Cells.Front Immunol. 2018 Aug 2;9:1785. doi: 10.3389/fimmu.2018.01785. eCollection 2018. Front Immunol. 2018. PMID: 30116248 Free PMC article.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials

