Stepwise mechanism for transcription fidelity
- PMID: 20459653
- PMCID: PMC2874521
- DOI: 10.1186/1741-7007-8-54
Stepwise mechanism for transcription fidelity
Abstract
Background: Transcription is the first step of gene expression and is characterized by a high fidelity of RNA synthesis. During transcription, the RNA polymerase active centre discriminates against not just non-complementary ribo NTP substrates but also against complementary 2'- and 3'-deoxy NTPs. A flexible domain of the RNA polymerase active centre, the Trigger Loop, was shown to play an important role in this process, but the mechanisms of this participation remained elusive.
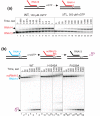
Results: Here we show that transcription fidelity is achieved through a multi-step process. The initial binding in the active centre is the major discrimination step for some non-complementary substrates, although for the rest of misincorporation events discrimination at this step is very poor. During the second step, non-complementary and 2'-deoxy NTPs are discriminated against based on differences in reaction transition state stabilization and partly in general base catalysis, for correct versus non-correct substrates. This step is determined by two residues of the Trigger Loop that participate in catalysis. In the following step, non-complementary and 2'-deoxy NTPs are actively removed from the active centre through a rearrangement of the Trigger Loop. The only step of discrimination against 3'-deoxy substrates, distinct from the ones above, is based on failure to orient the Trigger Loop catalytic residues in the absence of 3'OH.
Conclusions: We demonstrate that fidelity of transcription by multi-subunit RNA polymerases is achieved through a stepwise process. We show that individual steps contribute differently to discrimination against various erroneous substrates. We define the mechanisms and contributions of each of these steps to the overall fidelity of transcription.
Figures




References
-
- Sosunov V, Zorov S, Sosunova E, Nikolaev A, Zakeyeva I, Bass I, Goldfarb A, Nikiforov V, Severinov K, Mustaev A. The involvement of the aspartate triad of the active center in all catalytic activities of multisubunit RNA polymerase. Nucleic Acids Res. 2005;33:4202–4211. doi: 10.1093/nar/gki688. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- 202994/ERC_/European Research Council/International
- GM64530/GM/NIGMS NIH HHS/United States
- BB/I004564/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/F013558/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/F006462/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
LinkOut - more resources
Full Text Sources

