The Dunaliella salina organelle genomes: large sequences, inflated with intronic and intergenic DNA
- PMID: 20459666
- PMCID: PMC3017802
- DOI: 10.1186/1471-2229-10-83
The Dunaliella salina organelle genomes: large sequences, inflated with intronic and intergenic DNA
Abstract
Background: Dunaliella salina Teodoresco, a unicellular, halophilic green alga belonging to the Chlorophyceae, is among the most industrially important microalgae. This is because D. salina can produce massive amounts of beta-carotene, which can be collected for commercial purposes, and because of its potential as a feedstock for biofuels production. Although the biochemistry and physiology of D. salina have been studied in great detail, virtually nothing is known about the genomes it carries, especially those within its mitochondrion and plastid. This study presents the complete mitochondrial and plastid genome sequences of D. salina and compares them with those of the model green algae Chlamydomonas reinhardtii and Volvox carteri.
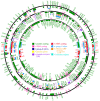
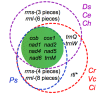
Results: The D. salina organelle genomes are large, circular-mapping molecules with approximately 60% noncoding DNA, placing them among the most inflated organelle DNAs sampled from the Chlorophyta. In fact, the D. salina plastid genome, at 269 kb, is the largest complete plastid DNA (ptDNA) sequence currently deposited in GenBank, and both the mitochondrial and plastid genomes have unprecedentedly high intron densities for organelle DNA: approximately 1.5 and approximately 0.4 introns per gene, respectively. Moreover, what appear to be the relics of genes, introns, and intronic open reading frames are found scattered throughout the intergenic ptDNA regions -- a trait without parallel in other characterized organelle genomes and one that gives insight into the mechanisms and modes of expansion of the D. salina ptDNA.
Conclusions: These findings confirm the notion that chlamydomonadalean algae have some of the most extreme organelle genomes of all eukaryotes. They also suggest that the events giving rise to the expanded ptDNA architecture of D. salina and other Chlamydomonadales may have occurred early in the evolution of this lineage. Although interesting from a genome evolution standpoint, the D. salina organelle DNA sequences will aid in the development of a viable plastid transformation system for this model alga, and they will complement the forthcoming D. salina nuclear genome sequence, placing D. salina in a group of a select few photosynthetic eukaryotes for which complete genome sequences from all three genetic compartments are available.
Figures



References
-
- Teodoresco EC. Organisation et développement du Dunaliella, nouveau genre de Volvocacée-Polyblépharidée. Beih Bot Zentralblatt Bd 18 Abt. 1905;1:215–232.
-
- Polle JEW, Tran D, Ben-Amotz A. In: The Alga Dunaliella: Biodiversity, Physiology, Genomics, and Biotechnology. Ben-Amotz A, Polle JEW, Rao DVS, editor. Enfield, New Hampshire, USA: Science Publishers; 2009. History, distribution, and habitats of algae of the genus Dunaliella Teodoresco (Chlorophyceae) pp. 1–14.
-
- Ben-Amotz A, Katz A, Avron M. Accumulation of β-carotene in halotolerant algae: Purification and characterization of β-carotene-rich globules from Dunaliella bardawil (Chlorophyceae) J Phycol. 1982;18:529–537. doi: 10.1111/j.1529-8817.1982.tb03219.x. - DOI
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

