Transcript profiling of common bean (Phaseolus vulgaris L.) using the GeneChip Soybean Genome Array: optimizing analysis by masking biased probes
- PMID: 20459672
- PMCID: PMC3017814
- DOI: 10.1186/1471-2229-10-85
Transcript profiling of common bean (Phaseolus vulgaris L.) using the GeneChip Soybean Genome Array: optimizing analysis by masking biased probes
Abstract
Background: Common bean (Phaseolus vulgaris L.) and soybean (Glycine max) both belong to the Phaseoleae tribe and share significant coding sequence homology. This suggests that the GeneChip(R) Soybean Genome Array (soybean GeneChip) may be used for gene expression studies using common bean.
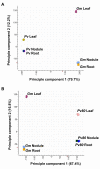
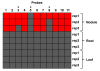
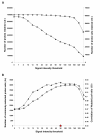
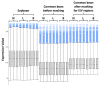
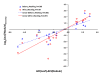
Results: To evaluate the utility of the soybean GeneChip for transcript profiling of common bean, we hybridized cRNAs purified from nodule, leaf, and root of common bean and soybean in triplicate to the soybean GeneChip. Initial data analysis showed a decreased sensitivity and accuracy of measuring differential gene expression in common bean cross-species hybridization (CSH) GeneChip data compared to that of soybean. We employed a method that masked putative probes targeting inter-species variable (ISV) regions between common bean and soybean. A masking signal intensity threshold was selected that optimized both sensitivity and accuracy of measuring differential gene expression. After masking for ISV regions, the number of differentially-expressed genes identified in common bean was increased by 2.8-fold reflecting increased sensitivity. Quantitative RT-PCR (qRT-PCR) analysis of 20 randomly selected genes and purine-ureide pathway genes demonstrated an increased accuracy of measuring differential gene expression after masking for ISV regions. We also evaluated masked probe frequency per probe set to gain insight into the sequence divergence pattern between common bean and soybean. The sequence divergence pattern analysis suggested that the genes for basic cellular functions and metabolism were highly conserved between soybean and common bean. Additionally, our results show that some classes of genes, particularly those associated with environmental adaptation, are highly divergent.
Conclusions: The soybean GeneChip is a suitable cross-species platform for transcript profiling in common bean when used in combination with the masking protocol described. In addition to transcript profiling, CSH of the GeneChip in combination with masking probes in the ISV regions can be used for comparative ecological and/or evolutionary genomics studies.
Figures

References
-
- Gepts P. Origin and evolution of common bean: past events and recent trends. Hort Sci. 1988;33:1124–1130.
-
- Food and Agriculture Organization of the United Nations. http://faostat.fao.org/
-
- McClean P, Kami J, Gepts P. In: Legume Crop Genomics. Wilson RF, Stalker HT, Brummer EC, editor. Champaign, IL: AOCS Press; 2004. Genomic and genetic diversity in common bean; pp. 60–82.
-
- Broughton WJ, Hernández G, Blair M, Beebe S, Gepts P, Vanderleyden J. Beans (Phaseolus spp.)-model food legume. Plant Soil. 2003;252:55–128. doi: 10.1023/A:1024146710611. - DOI
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

