The trans-Saharan slave trade - clues from interpolation analyses and high-resolution characterization of mitochondrial DNA lineages
- PMID: 20459715
- PMCID: PMC2875235
- DOI: 10.1186/1471-2148-10-138
The trans-Saharan slave trade - clues from interpolation analyses and high-resolution characterization of mitochondrial DNA lineages
Abstract
Background: A proportion of 1/4 to 1/2 of North African female pool is made of typical sub-Saharan lineages, in higher frequencies as geographic proximity to sub-Saharan Africa increases. The Sahara was a strong geographical barrier against gene flow, at least since 5,000 years ago, when desertification affected a larger region, but the Arab trans-Saharan slave trade could have facilitate enormously this migration of lineages. Till now, the genetic consequences of these forced trans-Saharan movements of people have not been ascertained.
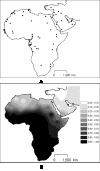
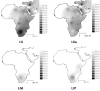
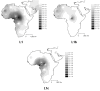
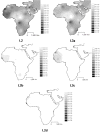
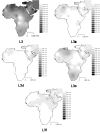
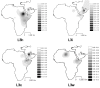
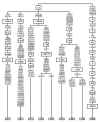
Results: The distribution of the main L haplogroups in North Africa clearly reflects the known trans-Saharan slave routes: West is dominated by L1b, L2b, L2c, L2d, L3b and L3d; the Center by L3e and some L3f and L3w; the East by L0a, L3h, L3i, L3x and, in common with the Center, L3f and L3w; while, L2a is almost everywhere. Ages for the haplogroups observed in both sides of the Saharan desert testify the recent origin (holocenic) of these haplogroups in sub-Saharan Africa, claiming a recent introduction in North Africa, further strengthened by the no detection of local expansions.
Conclusions: The interpolation analyses and complete sequencing of present mtDNA sub-Saharan lineages observed in North Africa support the genetic impact of recent trans-Saharan migrations, namely the slave trade initiated by the Arab conquest of North Africa in the seventh century. Sub-Saharan people did not leave traces in the North African maternal gene pool for the time of its settlement, some 40,000 years ago.
Figures


Similar articles
-
The genomic analysis of current-day North African populations reveals the existence of trans-Saharan migrations with different origins and dates.Hum Genet. 2023 Feb;142(2):305-320. doi: 10.1007/s00439-022-02503-3. Epub 2022 Nov 28. Hum Genet. 2023. PMID: 36441222 Free PMC article.
-
Ancient local evolution of African mtDNA haplogroups in Tunisian Berber populations.Hum Biol. 2010 Aug;82(4):367-84. doi: 10.3378/027.082.0402. Hum Biol. 2010. PMID: 21082907
-
Mitochondrial DNA diversity in the African American population.Mitochondrial DNA. 2015 Jun;26(3):445-51. doi: 10.3109/19401736.2013.840591. Epub 2013 Oct 9. Mitochondrial DNA. 2015. PMID: 24102597 Free PMC article.
-
African human mtDNA phylogeography at-a-glance.J Anthropol Sci. 2011;89:25-58. doi: 10.4436/jass.89006. Epub 2011 Mar 15. J Anthropol Sci. 2011. PMID: 21368343 Review.
-
A population genetics perspective of the Indus Valley through uniparentally-inherited markers.Ann Hum Biol. 2005 Mar-Apr;32(2):154-62. doi: 10.1080/03014460500076223. Ann Hum Biol. 2005. PMID: 16096211 Review.
Cited by
-
Genetic stratigraphy of key demographic events in Arabia.PLoS One. 2015 Mar 4;10(3):e0118625. doi: 10.1371/journal.pone.0118625. eCollection 2015. PLoS One. 2015. PMID: 25738654 Free PMC article.
-
The genomic analysis of current-day North African populations reveals the existence of trans-Saharan migrations with different origins and dates.Hum Genet. 2023 Feb;142(2):305-320. doi: 10.1007/s00439-022-02503-3. Epub 2022 Nov 28. Hum Genet. 2023. PMID: 36441222 Free PMC article.
-
Mitochondrial DNA and the Y chromosome suggest the settlement of Madagascar by Indonesian sea nomad populations.BMC Genomics. 2015 Mar 17;16(1):191. doi: 10.1186/s12864-015-1394-7. BMC Genomics. 2015. PMID: 25880430 Free PMC article.
-
Mycobacterium tuberculosis complex in remains of 18th-19th century slaves, Brazil.Emerg Infect Dis. 2013 May;19(5):837-9. doi: 10.3201/eid1905.120193. Emerg Infect Dis. 2013. PMID: 23697340 Free PMC article. No abstract available.
-
Tracing Arab-Islamic inheritance in Madagascar: study of the Y-chromosome and mitochondrial DNA in the Antemoro.PLoS One. 2013 Nov 22;8(11):e80932. doi: 10.1371/journal.pone.0080932. eCollection 2013. PLoS One. 2013. PMID: 24278350 Free PMC article.
References
-
- Behar DM, Metspalu E, Kivisild T, Achilli A, Hadid Y, Tzur S, Pereira L, Amorim A, Quintana-Murci L, Majamaa K, Herrnstadt C, Howell N, Balanovsky O, Kutuev I, Pshenichnov A, Gurwitz D, Bonne-Tamir B, Torroni A, Villems R, Skorecki K. The matrilineal ancestry of Ashkenazi Jewry: portrait of a recent founder event. Am J Hum Genet. 2006;78:487–497. doi: 10.1086/500307. - DOI - PMC - PubMed
-
- Behar DM, Metspalu E, Kivisild T, Rosset S, Tzur S, Hadid Y, Yudkovsky G, Rosengarten D, Pereira L, Amorim A, Kutuev I, Gurwitz D, Bonne-Tamir B, Villems R, Skorecki K. Counting the founders: the matrilineal genetic ancestry of the Jewish Diaspora. PLoS One. 2008;3:e2062. doi: 10.1371/journal.pone.0002062. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources

