Live cell visualization of the interactions between HIV-1 Gag and the cellular RNA-binding protein Staufen1
- PMID: 20459747
- PMCID: PMC2887380
- DOI: 10.1186/1742-4690-7-41
Live cell visualization of the interactions between HIV-1 Gag and the cellular RNA-binding protein Staufen1
Abstract
Background: Human immunodeficiency virus type 1 (HIV-1) uses cellular proteins and machinery to ensure transmission to uninfected cells. Although the host proteins involved in the transport of viral components toward the plasma membrane have been investigated, the dynamics of this process remain incompletely described. Previously we showed that the double-stranded (ds)RNA-binding protein, Staufen1 is found in the HIV-1 ribonucleoprotein (RNP) that contains the HIV-1 genomic RNA (vRNA), Gag and other host RNA-binding proteins in HIV-1-producing cells. Staufen1 interacts with the nucleocapsid domain (NC) domain of Gag and regulates Gag multimerization on membranes thereby modulating HIV-1 assembly. The formation of the HIV-1 RNP is dynamic and likely central to the fate of the vRNA during the late phase of the HIV-1 replication cycle.
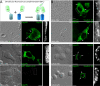
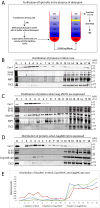
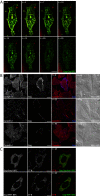
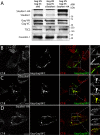
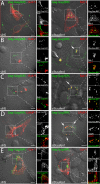
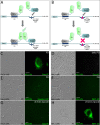
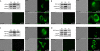
Results: Detailed molecular imaging of both the intracellular trafficking of virus components and of virus-host protein complexes is critical to enhance our understanding of factors that contribute to HIV-1 pathogenesis. In this work, we visualized the interactions between Gag and host proteins using bimolecular and trimolecular fluorescence complementation (BiFC and TriFC) analyses. These methods allow for the direct visualization of the localization of protein-protein and protein-protein-RNA interactions in live cells. We identified where the virus-host interactions between Gag and Staufen1 and Gag and IMP1 (also known as VICKZ1, IGF2BP1 and ZBP1) occur in cells. These virus-host interactions were not only detected in the cytoplasm, but were also found at cholesterol-enriched GM1-containing lipid raft plasma membrane domains. Importantly, Gag specifically recruited Staufen1 to the detergent insoluble membranes supporting a key function for this host factor during virus assembly. Notably, the TriFC experiments showed that Gag and Staufen1 actively recruited protein partners when tethered to mRNA.
Conclusions: The present work characterizes the interaction sites of key components of the HIV-1 RNP (Gag, Staufen1 and IMP1), thereby bringing to light where HIV-1 recruits and co-opts RNA-binding proteins during virus assembly.
Figures

Similar articles
-
How HIV-1 Gag Manipulates Its Host Cell Proteins: A Focus on Interactors of the Nucleocapsid Domain.Viruses. 2020 Aug 13;12(8):888. doi: 10.3390/v12080888. Viruses. 2020. PMID: 32823718 Free PMC article. Review.
-
Novel Staufen1 ribonucleoproteins prevent formation of stress granules but favour encapsidation of HIV-1 genomic RNA.J Cell Sci. 2010 Feb 1;123(Pt 3):369-83. doi: 10.1242/jcs.055897. Epub 2010 Jan 5. J Cell Sci. 2010. PMID: 20053637
-
HIV-1 NC-induced stress granule assembly and translation arrest are inhibited by the dsRNA binding protein Staufen1.RNA. 2018 Feb;24(2):219-236. doi: 10.1261/rna.064618.117. Epub 2017 Nov 10. RNA. 2018. PMID: 29127210 Free PMC article.
-
Characterization of staufen1 ribonucleoproteins by mass spectrometry and biochemical analyses reveal the presence of diverse host proteins associated with human immunodeficiency virus type 1.Front Microbiol. 2012 Oct 25;3:367. doi: 10.3389/fmicb.2012.00367. eCollection 2012. Front Microbiol. 2012. PMID: 23125841 Free PMC article.
-
Role of the nucleocapsid region in HIV-1 Gag assembly as investigated by quantitative fluorescence-based microscopy.Virus Res. 2014 Nov 26;193:78-88. doi: 10.1016/j.virusres.2014.06.009. Epub 2014 Jul 9. Virus Res. 2014. PMID: 25016037 Review.
Cited by
-
Staufen-1 interacts with the human endogenous retrovirus family HERV-K(HML-2) rec and gag proteins and increases virion production.J Virol. 2013 Oct;87(20):11019-30. doi: 10.1128/JVI.03031-12. Epub 2013 Aug 7. J Virol. 2013. PMID: 23926355 Free PMC article.
-
Analysis of the initiating events in HIV-1 particle assembly and genome packaging.PLoS Pathog. 2010 Nov 18;6(11):e1001200. doi: 10.1371/journal.ppat.1001200. PLoS Pathog. 2010. PMID: 21124996 Free PMC article.
-
The DEAD-box helicase DDX3 substitutes for the cap-binding protein eIF4E to promote compartmentalized translation initiation of the HIV-1 genomic RNA.Nucleic Acids Res. 2013 Jul;41(12):6286-99. doi: 10.1093/nar/gkt306. Epub 2013 Apr 28. Nucleic Acids Res. 2013. PMID: 23630313 Free PMC article.
-
Application of live-cell RNA imaging techniques to the study of retroviral RNA trafficking.Viruses. 2012 Jun;4(6):963-79. doi: 10.3390/v4060963. Epub 2012 Jun 8. Viruses. 2012. PMID: 22816035 Free PMC article. Review.
-
How HIV-1 Gag Manipulates Its Host Cell Proteins: A Focus on Interactors of the Nucleocapsid Domain.Viruses. 2020 Aug 13;12(8):888. doi: 10.3390/v12080888. Viruses. 2020. PMID: 32823718 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

