Vibrio chromosomes share common history
- PMID: 20459749
- PMCID: PMC2875227
- DOI: 10.1186/1471-2180-10-137
Vibrio chromosomes share common history
Abstract
Background: While most gamma proteobacteria have a single circular chromosome, Vibrionales have two circular chromosomes. Horizontal gene transfer is common among Vibrios, and in light of this genetic mobility, it is an open question to what extent the two chromosomes themselves share a common history since their formation.
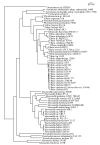
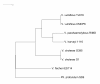
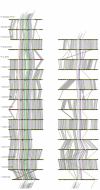
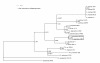
Results: Single copy genes from each chromosome (142 genes from chromosome I and 42 genes from chromosome II) were identified from 19 sequenced Vibrionales genomes and their phylogenetic comparison suggests consistent phylogenies for each chromosome. Additionally, study of the gene organization and phylogeny of the respective origins of replication confirmed the shared history.
Conclusions: Thus, while elements within the chromosomes may have experienced significant genetic mobility, the backbones share a common history. This allows conclusions based on multilocus sequence analysis (MLSA) for one chromosome to be applied equally to both chromosomes.
Figures
References
-
- Heidelberg JF, Eisen JA, Nelson WC, Clayton RA, Gwinn ML, Dodson RJ, Haft DH, Hickey EK, Peterson JD, Umayam L, Gill SR, Nelson KE, Read TD, Tettelin H, Richardson H, Ermolaeva MD, Vamathevan J, Bass S, Qin H, Dragoi I, Sellers P, McDonald L, Utterback T, Fleishmann RD, Nierman WC, White O, Salzberg SL, Smith HO, Colwell RR, Mekalanos JJ, Venter JC, Fraser CM. DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature. 2000;406:477–483. doi: 10.1038/35020000. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

