Interplay between Foxd3 and Mitf regulates cell fate plasticity in the zebrafish neural crest
- PMID: 20460180
- PMCID: PMC2909359
- DOI: 10.1016/j.ydbio.2010.04.023
Interplay between Foxd3 and Mitf regulates cell fate plasticity in the zebrafish neural crest
Abstract
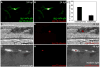
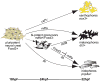
Pigment cells of the zebrafish, Danio rerio, offer an exceptionally tractable system for studying the genetic and cellular bases of cell fate decisions. In the zebrafish, neural crest cells generate three types of pigment cells during embryogenesis: yellow xanthophores, iridescent iridophores and black melanophores. In this study, we present evidence for a model whereby melanophores and iridophores descend from a common precursor whose fate is regulated by an interplay between the transcription factors Mitf and Foxd3. Loss of mitfa, a key regulator of melanophore development, resulted in supernumerary ectopic iridophores while loss of foxd3, a mitfa repressor, resulted in fewer iridophores. Double mutants showed a restoration of iridophores, suggesting that one of Foxd3's roles is to suppress mitfa to promote iridophore development. Foxd3 co-localized with pnp4a, a novel marker of early iridophore development, and was necessary for its expression. A considerable overlap was found between iridoblast and melanoblast markers but not xanthoblast markers, which resolved as cells began to differentiate. Cell lineage analyses using the photoconvertible marker, EosFP, revealed that both melanophores and iridophores develop from a mitfa+ precursor. Taken together, our data reveal a Foxd3/mitfa transcriptional switch that governs whether a bi-potent pigment precursor will attain either an iridophore or a melanophore fate.
Copyright (c) 2010 Elsevier Inc. All rights reserved.
Figures






References
-
- Adameyko I, Lallemend F, Aquino JB, Pereira JA, Topilko P, Muller T, Fritz N, Beljajeva A, Mochii M, Liste I, Usoskin D, Suter U, Birchmeier C, Ernfors P. Schwann cell precursors from nerve innervation are a cellular origin of melanocytes in skin. Cell. 2009;139:366–79. - PubMed
-
- Bagnara JT, Fernandez PJ, Fujii R. On the blue coloration of vertebrates. Pigment Cell Res. 2007;20:14–26. - PubMed
-
- Bagnara JT, Matsumoto J, Ferris W, Frost SK, Turner WA, Jr, Tchen TT, Taylor JD. Common origin of pigment cells. Science. 1979;203:410–5. - PubMed
-
- Bondurand N, Pingault V, Goerich DE, Lemort N, Sock E, Le Caignec C, Wegner M, Goossens M. Interaction among SOX10, PAX3 and MITF, three genes altered in Waardenburg syndrome. Hum Mol Genet. 2000;9:1907–17. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

