The non-coding RNA TERRA is a natural ligand and direct inhibitor of human telomerase
- PMID: 20460456
- PMCID: PMC2943627
- DOI: 10.1093/nar/gkq296
The non-coding RNA TERRA is a natural ligand and direct inhibitor of human telomerase
Abstract
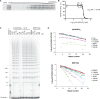
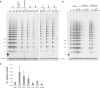
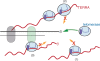
Telomeres, the physical ends of eukaryotes chromosomes are transcribed into telomeric repeat containing RNA (TERRA), a large non-coding RNA of unknown function, which forms an integral part of telomeric heterochromatin. TERRA molecules resemble in sequence the telomeric DNA substrate as they contain 5'-UUAGGG-3' repeats near their 3'-end which are complementary to the template sequence of telomerase RNA. Here we demonstrate that endogenous TERRA is bound to human telomerase in cell extracts. Using in vitro reconstituted telomerase and synthetic TERRA molecules we demonstrate that the 5'-UUAGGG-3' repeats of TERRA base pair with the RNA template of the telomerase RNA moiety (TR). In addition TERRA contacts the telomerase reverse transcriptase (TERT) protein subunit independently of hTR. In vitro studies further demonstrate that TERRA is not used as a telomerase substrate. Instead, TERRA acts as a potent competitive inhibitor for telomeric DNA in addition to exerting an uncompetitive mode of inhibition. Our data identify TERRA as a telomerase ligand and natural direct inhibitor of human telomerase. Telomerase regulation by the telomere substrate may be mediated via its transcription.
Figures



References
-
- Greider CW, Blackburn EH. Identification of a specific telomere terminal transferase activity in Tetrahymena extracts. Cell. 1985;43:405–413. - PubMed
-
- Cech TR. Beginning to understand the end of the chromosome. Cell. 2004;116:273–279. - PubMed
-
- Ottaviani A, Gilson E, Magdinier F. Telomeric position effect: from the yeast paradigm to human pathologies? Biochimie. 2008;90:93–107. - PubMed
-
- Blasco MA. The epigenetic regulation of mammalian telomeres. Nat. Rev. Genet. 2007;8:299–309. - PubMed

