Functional modularity of nuclear hormone receptors in a Caenorhabditis elegans metabolic gene regulatory network
- PMID: 20461074
- PMCID: PMC2890327
- DOI: 10.1038/msb.2010.23
Functional modularity of nuclear hormone receptors in a Caenorhabditis elegans metabolic gene regulatory network
Abstract
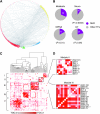
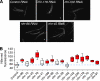
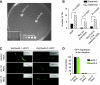
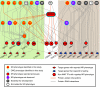
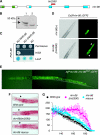
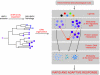
Gene regulatory networks (GRNs) provide insights into the mechanisms of differential gene expression at a systems level. GRNs that relate to metazoan development have been studied extensively. However, little is still known about the design principles, organization and functionality of GRNs that control physiological processes such as metabolism, homeostasis and responses to environmental cues. In this study, we report the first experimentally mapped metazoan GRN of Caenorhabditis elegans metabolic genes. This network is enriched for nuclear hormone receptors (NHRs). The NHR family has greatly expanded in nematodes: humans have 48 NHRs, but C. elegans has 284, most of which are uncharacterized. We find that the C. elegans metabolic GRN is highly modular and that two GRN modules predominantly consist of NHRs. Network modularity has been proposed to facilitate a rapid response to different cues. As NHRs are metabolic sensors that are poised to respond to ligands, this suggests that C. elegans GRNs evolved to enable rapid and adaptive responses to different cues by a concurrence of NHR family expansion and modular GRN wiring.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures


References
-
- Alon U (2007) Network motifs: theory and experimental approaches. Nat Rev Genet 8: 450–461 - PubMed
-
- Ashrafi K, Chang FY, Watts JL, Fraser AG, Kamath RS, Ahringer J, Ruvkun G (2003) Genome-wide RNAi analysis of Caenorhabditis elegans fat regulatory genes. Nature 421: 268–272 - PubMed
-
- Babu MM, Luscombe NM, Aravind L, Gerstein M, Teichmann SA (2004) Structure and evolution of transcriptional regulatory networks. Curr Opin Struct Biol 14: 283–291 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

