A general lack of compensation for gene dosage in yeast
- PMID: 20461075
- PMCID: PMC2890323
- DOI: 10.1038/msb.2010.19
A general lack of compensation for gene dosage in yeast
Abstract
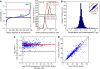
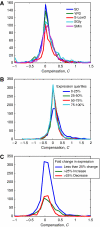
Gene copy number variation has been discovered in humans, between related species, and in different cancer tissues, but it is unclear how much of this genomic-level variation leads to changes in the level of protein abundance. To address this, we eliminated one of the two genomic copies of 730 different genes in Saccharomyces cerevisiae and asked how often a 50% reduction in gene dosage leads to a 50% reduction in protein level. For at least 80% of genes tested, and under several environmental conditions, it does: protein levels in the heterozygous strain are close to 50% of wild type. For <5% of the genes tested, the protein levels in the heterozygote are maintained at nearly wild-type levels. These experiments show that protein levels are not, in general, directly monitored and adjusted to a desired level. Combined with fitness data, this implies that proteins are expressed at levels higher than necessary for survival.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures

Comment in
-
Functional genomics: One gene or two?Nat Rev Genet. 2010 Jul;11(7):453. doi: 10.1038/nrg2816. Nat Rev Genet. 2010. PMID: 20548293 No abstract available.
References
-
- Brooker RJ, Widmaier EP, Graham LE, Stiling PD (2008) Biology, 1st edn. McGraw Hill, Boston, MA, USA
-
- Cleveland DW, Lopata MA, Sherline P, Kirschner MW (1981) Unpolymerized tubulin modulates the level of tubulin mRNAs. Cell 25: 537–546 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

