Actin dynamics: from nanoscale to microscale
- PMID: 20462375
- PMCID: PMC2967719
- DOI: 10.1146/annurev.biophys.093008.131207
Actin dynamics: from nanoscale to microscale
Abstract
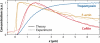
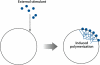
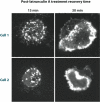
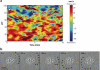
The dynamic nature of actin in cells manifests itself constantly. Polymerization near the cell edge is balanced by depolymerization in the interior, externally induced actin polymerization is followed by depolymerization, and spontaneous oscillations of actin at the cell periphery are frequently seen. I discuss how mathematical modeling relates quantitative measures of actin dynamics to the rates of underlying molecular level processes. The dynamic properties addressed include the rate of actin assembly at the leading edge of a moving cell, the disassembly rates of intracellular actin networks, the polymerization time course in externally stimulated cells, and spontaneous spatiotemporal patterns formed by actin. Although several aspects of actin assembly have been clarified by increasingly sophisticated models, our understanding of rapid actin disassembly is limited, and the origins of nonmonotonic features in externally stimulated actin polymerization remain unclear. Theory has generated several concrete, testable hypotheses for the origins of spontaneous actin waves and cell-edge oscillations. The development and use of more biomimetic systems applicable to the geometry of a cell will be key to obtaining a quantitative understanding of actin dynamics in cells.
Figures



References
-
- Alon U. An Introduction to Systems Biology: Design Principles of Biological Circuits. Chapman & Hall/CRC; Boca Raton, FL: 2007. pp. 41–73. Chapter 4: “The Feedforward Network Loop Motif”.
-
- Betz T, Koch D, Lim D, Käs JA. Stochastic actin polymerization and steady retrograde flow determine growth cone advancement. Biophys. J. 2009;96:5130–-38. [5. Used a combination of microscopy and quantitative statistical analysis to establish strong correlations between cell-edge oscillations and actin polymerization] - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

