Nucleosome rotational setting is associated with transcriptional regulation in promoters of tissue-specific human genes
- PMID: 20462404
- PMCID: PMC2898081
- DOI: 10.1186/gb-2010-11-5-r51
Nucleosome rotational setting is associated with transcriptional regulation in promoters of tissue-specific human genes
Abstract
Background: The position of a nucleosome, both translational along the DNA molecule and rotational between the histone core and the DNA, is controlled by many factors, including the regular occurrence of specific dinucleotides with a period of approximately 10 bp, important for the rotational setting of the DNA around the histone octamer.
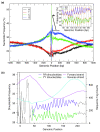
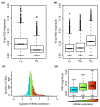
Results: We show that such a 10 bp periodic signal of purine-purine dinucleotides occurs in phase with the transcription start site (TSS) of human genes and is centered on the position of the first (+1) nucleosome downstream of the TSS. These data support a direct link between transcription and the rotational setting of the nucleosome. The periodic signal is most prevalent in genes that contain CpG islands that are expressed at low levels in a tissue-specific manner and are involved in the control of transcription.
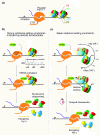
Conclusions: These results, together with several lines of evidence from the recent literature, support a new model whereby the +1 nucleosome could be more efficiently disassembled from gene promoters by H3K56 acetylation marks if the periodic signal specifies an optimal rotational setting.
Figures


Similar articles
-
Influence of Rotational Nucleosome Positioning on Transcription Start Site Selection in Animal Promoters.PLoS Comput Biol. 2016 Oct 7;12(10):e1005144. doi: 10.1371/journal.pcbi.1005144. eCollection 2016 Oct. PLoS Comput Biol. 2016. PMID: 27716823 Free PMC article.
-
Histone H3 acetylated at lysine 9 in promoter is associated with low nucleosome density in the vicinity of transcription start site in human cell.Chromosome Res. 2006;14(2):203-11. doi: 10.1007/s10577-006-1036-7. Epub 2006 Mar 17. Chromosome Res. 2006. PMID: 16544193
-
Contrasting chromatin organization of CpG islands and exons in the human genome.Genome Biol. 2010;11(7):R70. doi: 10.1186/gb-2010-11-7-r70. Epub 2010 Jul 5. Genome Biol. 2010. PMID: 20602769 Free PMC article.
-
Interplay between the transcription preinitiation complex and the +1 nucleosome.Trends Biochem Sci. 2024 Feb;49(2):145-155. doi: 10.1016/j.tibs.2023.12.001. Epub 2024 Jan 13. Trends Biochem Sci. 2024. PMID: 38218671 Review.
-
Nucleosome positioning in yeasts: methods, maps, and mechanisms.Chromosoma. 2015 Jun;124(2):131-51. doi: 10.1007/s00412-014-0501-x. Epub 2014 Dec 23. Chromosoma. 2015. PMID: 25529773 Review.
Cited by
-
Codons support the maintenance of intrinsic DNA polymer flexibility over evolutionary timescales.Genome Biol Evol. 2012;4(9):954-65. doi: 10.1093/gbe/evs073. Epub 2012 Aug 30. Genome Biol Evol. 2012. PMID: 22936074 Free PMC article.
-
Functional conservation of nucleosome formation selectively biases presumably neutral molecular variation in yeast genomes.Genome Biol Evol. 2011;3:15-22. doi: 10.1093/gbe/evq081. Epub 2010 Dec 6. Genome Biol Evol. 2011. PMID: 21135411 Free PMC article.
-
Oncogene Ras/phosphatidylinositol 3-kinase signaling targets histone H3 acetylation at lysine 56.J Biol Chem. 2012 Nov 30;287(49):41469-80. doi: 10.1074/jbc.M112.367847. Epub 2012 Sep 16. J Biol Chem. 2012. PMID: 22982396 Free PMC article.
-
The type III histone deacetylase Sirt1 protein suppresses p300-mediated histone H3 lysine 56 acetylation at Bclaf1 promoter to inhibit T cell activation.J Biol Chem. 2011 May 13;286(19):16967-75. doi: 10.1074/jbc.M111.218206. Epub 2011 Mar 22. J Biol Chem. 2011. PMID: 21454709 Free PMC article.
-
Apoptotic lymphocytes of H. sapiens lose nucleosomes in GC-rich promoters.PLoS Comput Biol. 2014 Jul 31;10(7):e1003760. doi: 10.1371/journal.pcbi.1003760. eCollection 2014 Jul. PLoS Comput Biol. 2014. PMID: 25077608 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

