Coordinated modular functionality and prognostic potential of a heart failure biomarker-driven interaction network
- PMID: 20462429
- PMCID: PMC2890499
- DOI: 10.1186/1752-0509-4-60
Coordinated modular functionality and prognostic potential of a heart failure biomarker-driven interaction network
Abstract
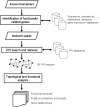
Background: The identification of potentially relevant biomarkers and a deeper understanding of molecular mechanisms related to heart failure (HF) development can be enhanced by the implementation of biological network-based analyses. To support these efforts, here we report a global network of protein-protein interactions (PPIs) relevant to HF, which was characterized through integrative bioinformatic analyses of multiple sources of "omic" information.
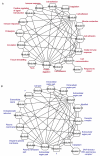
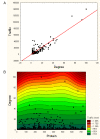
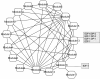
Results: We found that the structural and functional architecture of this PPI network is highly modular. These network modules can be assigned to specialized processes, specific cellular regions and their functional roles tend to partially overlap. Our results suggest that HF biomarkers may be defined as key coordinators of intra- and inter-module communication. Putative biomarkers can, in general, be distinguished as "information traffic" mediators within this network. The top high traffic proteins are encoded by genes that are not highly differentially expressed across HF and non-HF patients. Nevertheless, we present evidence that the integration of expression patterns from high traffic genes may support accurate prediction of HF. We quantitatively demonstrate that intra- and inter-module functional activity may be controlled by a family of transcription factors known to be associated with the prevention of hypertrophy.
Conclusion: The systems-driven analysis reported here provides the basis for the identification of potentially novel biomarkers and understanding HF-related mechanisms in a more comprehensive and integrated way.
Figures

References
-
- Rual JF, Venkatesan K, Hao T, Hirozane-Kishikawa T, Dricot A, Li N, Berriz GF, Gibbons FD, Dreze M, Ayivi-Guedehoussou N, Klitgord N, Simon C, Boxem M, Milstein S, Rosenberg J, Goldberg DS, Zhang LV, Wong SL, Franklin G, Li S, Albala JS, Lim J, Fraughton C, Llamosas E, Cevik S, Bex C, Lamesch P, Sikorski RS, Vandenhaute J, Zoghbi HY, Smolyar A, Bosak S, Sequerra R, Doucette-Stamm L, Cusick ME, Hill DE, Roth FP, Vidal M. Towards a proteome-scale map of the human protein-protein interaction network. Nature. 2005;437:1173–8. doi: 10.1038/nature04209. - DOI - PubMed
-
- Stelzl U, Worm U, Lalowski M, Haenig C, Brembeck FH, Goehler H, Stroedicke M, Zenkner M, Schoenherr A, Koeppen S, Timm J, Mintzlaff S, Abraham C, Bock N, Kietzmann S, Goedde A, Toksöz E, Droege A, Krobitsch S, Korn B, Birchmeier W, Lehrach H, Wanker EE. A human protein-protein interaction network: a resource for annotating the proteome. Cell. 2005;122:957–68. doi: 10.1016/j.cell.2005.08.029. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

