Smad2 and Smad6 as predictors of overall survival in oral squamous cell carcinoma patients
- PMID: 20462450
- PMCID: PMC2885344
- DOI: 10.1186/1476-4598-9-106
Smad2 and Smad6 as predictors of overall survival in oral squamous cell carcinoma patients
Abstract
Background: To test if the expression of Smad1-8 mRNAs were predictive of survival in patients with oral squamous cell carcinoma (SCC).
Patients and methods: We analyzed, prospectively, the expression of Smad1-8, by means of Ribonuclease Protection Assay in 48 primary, operable, oral SCC. In addition, 21 larynx, 10 oropharynx and 4 hypopharynx SCC and 65 matched adjacent mucosa, available for study, were also included. For survival analysis, patients were categorized as positive or negative for each Smad, according to median mRNA expression. We also performed real-time quantitative PCR (QRTPCR) to asses the pattern of TGFbeta1, TGFbeta2, TGFbeta3 in oral SCC.
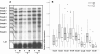
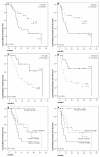
Results: Our results showed that Smad2 and Smad6 mRNA expression were both associated with survival in Oral SCC patients. Cox Multivariate analysis revealed that Smad6 positivity and Smad2 negativity were both predictive of good prognosis for oral SCC patients, independent of lymph nodal status (P = 0.003 and P = 0.029, respectively). In addition, simultaneously Smad2- and Smad6+ oral SCC group of patients did not reach median overall survival (mOS) whereas the mOS of Smad2+/Smad6- subgroup was 11.6 months (P = 0.004, univariate analysis). Regarding to TGFbeta isoforms, we found that Smad2 mRNA and TGFbeta1 mRNA were inversely correlated (p = 0.05, R = -0.33), and that seven of the eight TGFbeta1+ patients were Smad2-. In larynx SCC, Smad7- patients did not reach mOS whereas mOS of Smad7+ patients were only 7.0 months (P = 0.04). No other correlations were found among Smad expression, clinico-pathological characteristics and survival in oral, larynx, hypopharynx, oropharynx or the entire head and neck SCC population.
Conclusion: Smad6 together with Smad2 may be prognostic factors, independent of nodal status in oral SCC after curative resection. The underlying mechanism which involves aberrant TGFbeta signaling should be better clarified in the future.
Figures


Similar articles
-
Transforming growth factor beta1 (TGFbeta1) expression in head and neck squamous cell carcinoma patients as related to prognosis.J Oral Pathol Med. 2003 Mar;32(3):139-45. doi: 10.1034/j.1600-0714.2003.00012.x. J Oral Pathol Med. 2003. PMID: 12581383
-
Overexpression of Smad proteins, especially Smad7, in oral epithelial dysplasias.Clin Oral Investig. 2013 Apr;17(3):921-32. doi: 10.1007/s00784-012-0756-7. Epub 2012 Jun 6. Clin Oral Investig. 2013. PMID: 22669485
-
Downregulation of SMAD2, 4 and 6 mRNA and TGFbeta receptor I mRNA in lesional and non-lesional psoriatic skin.Acta Derm Venereol. 2009;89(4):351-6. doi: 10.2340/00015555-0634. Acta Derm Venereol. 2009. PMID: 19688145
-
Prognostic value of the expression of Smad6 and Smad7, as inhibitory Smads of the TGF-beta superfamily, in esophageal squamous cell carcinoma.Anticancer Res. 2004 Nov-Dec;24(6):3703-9. Anticancer Res. 2004. PMID: 15736400
-
Defective transforming growth factor beta signaling pathway in head and neck squamous cell carcinoma as evidenced by the lack of expression of activated Smad2.Clin Cancer Res. 2001 Jun;7(6):1618-26. Clin Cancer Res. 2001. PMID: 11410498
Cited by
-
Aberrations in SMAD family of genes among HNSCC patients.Bioinformation. 2021 Dec 31;17(12):113-119. doi: 10.6026/973206300171113. eCollection 2021. Bioinformation. 2021. PMID: 35291342 Free PMC article.
-
Identifying Novel Candidate Genes Related to Apoptosis from a Protein-Protein Interaction Network.Comput Math Methods Med. 2015;2015:715639. doi: 10.1155/2015/715639. Epub 2015 Oct 4. Comput Math Methods Med. 2015. PMID: 26543496 Free PMC article.
-
Berberine Suppresses EMT in Liver and Gastric Carcinoma Cells through Combination with TGFβR Regulating TGF-β/Smad Pathway.Oxid Med Cell Longev. 2021 Oct 18;2021:2337818. doi: 10.1155/2021/2337818. eCollection 2021. Oxid Med Cell Longev. 2021. PMID: 34712379 Free PMC article.
-
Highly preserved consensus gene modules in human papilloma virus 16 positive cervical cancer and head and neck cancers.Oncotarget. 2017 Dec 7;8(69):114031-114040. doi: 10.18632/oncotarget.23116. eCollection 2017 Dec 26. Oncotarget. 2017. PMID: 29371966 Free PMC article.
-
Head and Neck Squamous Cell Carcinoma Biopsies Maintained Ex Vivo on a Perfusion Device Show Gene Changes with Time and Clinically Relevant Doses of Irradiation.Cancers (Basel). 2023 Sep 15;15(18):4575. doi: 10.3390/cancers15184575. Cancers (Basel). 2023. PMID: 37760543 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

