Massively parallel pyrosequencing-based transcriptome analyses of small brown planthopper (Laodelphax striatellus), a vector insect transmitting rice stripe virus (RSV)
- PMID: 20462456
- PMCID: PMC2885366
- DOI: 10.1186/1471-2164-11-303
Massively parallel pyrosequencing-based transcriptome analyses of small brown planthopper (Laodelphax striatellus), a vector insect transmitting rice stripe virus (RSV)
Abstract
Background: The small brown planthopper (Laodelphax striatellus) is an important agricultural pest that not only damages rice plants by sap-sucking, but also acts as a vector that transmits rice stripe virus (RSV), which can cause even more serious yield loss. Despite being a model organism for studying entomology, population biology, plant protection, molecular interactions among plants, viruses and insects, only a few genomic sequences are available for this species. To investigate its transcriptome and determine the differences between viruliferous and naïve L. striatellus, we employed 454-FLX high-throughput pyrosequencing to generate EST databases of this insect.
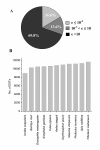
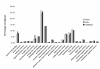
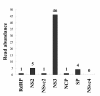
Results: We obtained 201,281 and 218,681 high-quality reads from viruliferous and naïve L. striatellus, respectively, with an average read length as 230 bp. These reads were assembled into contigs and two EST databases were generated. When all reads were combined, 16,885 contigs and 24,607 singletons (a total of 41,492 unigenes) were obtained, which represents a transcriptome of the insect. BlastX search against the NCBI-NR database revealed that only 6,873 (16.6%) of these unigenes have significant matches. Comparison of the distribution of GO classification among viruliferous, naïve, and combined EST databases indicated that these libraries are broadly representative of the L. striatellus transcriptomes. Functionally diverse transcripts from RSV, endosymbiotic bacteria Wolbachia and yeast-like symbiotes were identified, which reflects the possible lifestyles of these microbial symbionts that live in the cells of the host insect. Comparative genomic analysis revealed that L. striatellus encodes similar innate immunity regulatory systems as other insects, such as RNA interference, JAK/STAT and partial Imd cascades, which might be involved in defense against viral infection. In addition, we determined the differences in gene expression between vector and naïve samples, which generated a list of candidate genes that are potentially involved in the symbiosis of L. striatellus and RSV.
Conclusions: To our knowledge, the present study is the first description of a genomic project for L. striatellus. The identification of transcripts from RSV, Wolbachia, yeast-like symbiotes and genes abundantly expressed in viruliferous insect, provided a starting-point for investigating the molecular basis of symbiosis among these organisms.
Figures

Similar articles
-
Comparative Transcriptome Analysis of Chemoreception Organs of Laodelphax striatellus in Response to Rice Stripe Virus Infection.Int J Mol Sci. 2021 Sep 24;22(19):10299. doi: 10.3390/ijms221910299. Int J Mol Sci. 2021. PMID: 34638638 Free PMC article.
-
Alternative Splicing Landscape of Small Brown Planthopper and Different Response of JNK2 Isoforms to Rice Stripe Virus Infection.J Virol. 2022 Jan 26;96(2):e0171521. doi: 10.1128/JVI.01715-21. Epub 2021 Nov 10. J Virol. 2022. PMID: 34757837 Free PMC article.
-
The small brown planthopper (Laodelphax striatellus) as a vector of the rice stripe virus.Arch Insect Biochem Physiol. 2023 Feb;112(2):e21992. doi: 10.1002/arch.21992. Epub 2022 Dec 27. Arch Insect Biochem Physiol. 2023. PMID: 36575628 Review.
-
Rice stripe virus-derived siRNAs play different regulatory roles in rice and in the insect vector Laodelphax striatellus.BMC Plant Biol. 2018 Oct 4;18(1):219. doi: 10.1186/s12870-018-1438-7. BMC Plant Biol. 2018. PMID: 30286719 Free PMC article.
-
Migration of rice planthoppers and their vectored re-emerging and novel rice viruses in East Asia.Front Microbiol. 2013 Oct 28;4:309. doi: 10.3389/fmicb.2013.00309. Front Microbiol. 2013. PMID: 24312081 Free PMC article. Review.
Cited by
-
Rice stripe tenuivirus nonstructural protein 3 hijacks the 26S proteasome of the small brown planthopper via direct interaction with regulatory particle non-ATPase subunit 3.J Virol. 2015 Apr;89(8):4296-310. doi: 10.1128/JVI.03055-14. Epub 2015 Feb 4. J Virol. 2015. PMID: 25653432 Free PMC article.
-
Generation of a Transcriptome in a Model Lepidopteran Pest, Heliothis virescens, Using Multiple Sequencing Strategies for Profiling Midgut Gene Expression.PLoS One. 2015 Jun 5;10(6):e0128563. doi: 10.1371/journal.pone.0128563. eCollection 2015. PLoS One. 2015. PMID: 26047101 Free PMC article.
-
Population diversity of rice stripe virus-derived siRNAs in three different hosts and RNAi-based antiviral immunity in Laodelphgax striatellus.PLoS One. 2012;7(9):e46238. doi: 10.1371/journal.pone.0046238. Epub 2012 Sep 28. PLoS One. 2012. PMID: 23029445 Free PMC article.
-
Determination of Suitable RT-qPCR Reference Genes for Studies of Gene Functions in Laodelphax striatellus (Fallén).Genes (Basel). 2019 Nov 4;10(11):887. doi: 10.3390/genes10110887. Genes (Basel). 2019. PMID: 31689985 Free PMC article.
-
Transcriptome of the inflorescence meristems of the biofuel plant Jatropha curcas treated with cytokinin.BMC Genomics. 2014 Nov 17;15(1):974. doi: 10.1186/1471-2164-15-974. BMC Genomics. 2014. PMID: 25400171 Free PMC article.
References
-
- Toriyama S. Rice stripe virus: prototype of a new group of viruses that replicate in plants and insects. Microbiol Sci. 1986;3(11):347–351. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

