Physical basis of the inducer-dependent cooperativity of the Central glycolytic genes Repressor/DNA complex
- PMID: 20462860
- PMCID: PMC2943609
- DOI: 10.1093/nar/gkq334
Physical basis of the inducer-dependent cooperativity of the Central glycolytic genes Repressor/DNA complex
Abstract
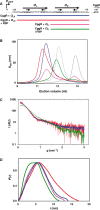
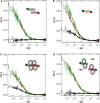
The Central glycolytic genes Repressor (CggR) from Bacillus subtilis belongs to the SorC family of transcription factors that control major carbohydrate metabolic pathways. Recent studies have shown that CggR binds as a tetramer to its tandem operator DNA sequences and that the inducer metabolite, fructose 1,6-bisphosphate (FBP), reduces the binding cooperativity of the CggR/DNA complex. Here, we have determined the effect of FBP on the size, shape and stoichiometry of CggR complexes with full-length and half-site operator sequence by small-angle X-ray scattering, size-exclusion chromatography, fluorescence cross-correlation spectroscopy and noncovalent mass spectrometry (MS). Our results show that CggR forms a compact tetrameric assembly upon binding to either the full-length operator or two half-site DNAs and that FBP triggers a tetramer-dimer transition that leaves a single dimer on the half-site or two physically independent dimers on the full-length target. Although the binding of other phospho-sugars was evidenced by MS, only FBP was found to completely disrupt dimer-dimer contacts. We conclude that inducer-dependent dimer-dimer bridging interactions constitute the physical basis for CggR cooperative binding to DNA and the underlying repression mechanism. This work provides experimental evidences for a cooperativity-based regulation model that should apply to other SorC family members.
Figures



References
-
- Shea MA, Ackers GK. The OR control system of bacteriophage lambda. A physical-chemical model for gene regulation. J. Mol. Biol. 1985;181:211–230. - PubMed
-
- Grillo AO, Brown MP, Royer CA. Probing the physical basis for trp repressor-operator recognition. J. Mol. Biol. 1999;287:539–554. - PubMed
-
- Chahla M, Wooll J, Laue TM, Nguyen N, Senear DF. Role of protein-protein bridging interactions on cooperative assembly of DNA-bound CRP-CytR-CRP complex and regulation of the Escherichia coli CytR regulon. Biochemistry. 2003;42:3812–3825. - PubMed
-
- Beckett D. Linked equilibria in regulation of transcription initiation. Methods Cell Biol. 2008;84:25–52. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

