Human cytochrome P450 2E1 structures with fatty acid analogs reveal a previously unobserved binding mode
- PMID: 20463018
- PMCID: PMC2903405
- DOI: 10.1074/jbc.M110.109017
Human cytochrome P450 2E1 structures with fatty acid analogs reveal a previously unobserved binding mode
Abstract
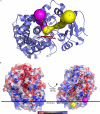
Human microsomal cytochrome P450 (CYP) 2E1 is widely known for its ability to oxidize >70 different, mostly compact, low molecular weight drugs and other xenobiotic compounds. In addition CYP2E1 oxidizes much larger C9-C20 fatty acids that can serve as endogenous signaling molecules. Previously structures of CYP2E1 with small molecules revealed a small, compact CYP2E1 active site, which would be insufficient to accommodate medium and long chain fatty acids without conformational changes in the protein. In the current work we have determined how CYP2E1 can accommodate a series of fatty acid analogs by cocrystallizing CYP2E1 with omega-imidazolyl-octanoic fatty acid, omega-imidazolyl-decanoic fatty acid, and omega-imidazolyl-dodecanoic fatty acid. In each structure direct coordination of the imidazole nitrogen to the heme iron mimics the position required for native fatty acid substrates to yield the omega-1 hydroxylated metabolites that predominate experimentally. In each case rotation of a single Phe(298) side chain merges the active site with an adjacent void, significantly altering the active site size and topology to accommodate fatty acids. The binding of these fatty acid ligands is directly opposite the channel to the protein surface and the binding observed for fatty acids in the bacterial cytochrome P450 BM3 (CYP102A1) from Bacillus megaterium. Instead of the BM3-like binding mode in the CYP2E1 channel, these structures reveal interactions between the fatty acid carboxylates and several residues in the F, G, and B' helices at successive distances from the active site.
Figures



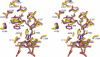

Similar articles
-
Structures of human cytochrome P-450 2E1. Insights into the binding of inhibitors and both small molecular weight and fatty acid substrates.J Biol Chem. 2008 Nov 28;283(48):33698-707. doi: 10.1074/jbc.M805999200. Epub 2008 Sep 24. J Biol Chem. 2008. PMID: 18818195 Free PMC article.
-
Imidazolyl carboxylic acids as mechanistic probes of flavocytochrome P-450 BM3.Biochemistry. 1998 Nov 10;37(45):15799-807. doi: 10.1021/bi980462d. Biochemistry. 1998. PMID: 9843385
-
[Computer modeling of cytochrome P450 2E1 three-dimensional structure].Ukr Biokhim Zh (1999). 2006 Mar-Apr;78(2):154-62. Ukr Biokhim Zh (1999). 2006. PMID: 17100298 Russian.
-
Negatively cooperative binding properties of human cytochrome P450 2E1 with monocyclic substrates.Curr Drug Metab. 2012 Sep 1;13(7):1024-31. doi: 10.2174/138920012802138606. Curr Drug Metab. 2012. PMID: 22591346 Review.
-
Advances in the interpretation and prediction of CYP2E1 metabolism from a biochemical perspective.Expert Opin Drug Metab Toxicol. 2008 Aug;4(8):1053-64. doi: 10.1517/17425255.4.8.1053. Expert Opin Drug Metab Toxicol. 2008. PMID: 18680440 Review.
Cited by
-
Human cytochrome P450 1A1 structure and utility in understanding drug and xenobiotic metabolism.J Biol Chem. 2013 May 3;288(18):12932-43. doi: 10.1074/jbc.M113.452953. Epub 2013 Mar 18. J Biol Chem. 2013. PMID: 23508959 Free PMC article.
-
Roles of Cytochrome P450 in Metabolism of Ethanol and Carcinogens.Adv Exp Med Biol. 2018;1032:15-35. doi: 10.1007/978-3-319-98788-0_2. Adv Exp Med Biol. 2018. PMID: 30362088 Free PMC article. Review.
-
Q11, a CYP2E1 inhibitor, exerts anti-hepatocellular carcinoma effect by inhibiting M2 macrophage polarization.Cancer Immunol Immunother. 2024 Dec 30;74(1):35. doi: 10.1007/s00262-024-03912-1. Cancer Immunol Immunother. 2024. PMID: 39738913 Free PMC article.
-
Kinetic characterizations of diallyl sulfide analogs for their novel role as CYP2E1 enzyme inhibitors.Pharmacol Res Perspect. 2017 Oct;5(5):e00362. doi: 10.1002/prp2.362. Pharmacol Res Perspect. 2017. PMID: 28971616 Free PMC article.
-
Crosstalk between CYP2E1 and PPARα substrates and agonists modulate adipose browning and obesity.Acta Pharm Sin B. 2022 May;12(5):2224-2238. doi: 10.1016/j.apsb.2022.02.004. Epub 2022 Feb 11. Acta Pharm Sin B. 2022. PMID: 35646522 Free PMC article.
References
-
- Laethem R. M., Balazy M., Falck J. R., Laethem C. L., Koop D. R. (1993) J. Biol. Chem. 268, 12912–12918 - PubMed
-
- Lucas D., Farez C., Bardou L. G., Vaisse J., Attali J. R., Valensi P. (1998) Fundam. Clin. Pharmacol. 12, 553–558 - PubMed
-
- McCarver D. G., Byun R., Hines R. N., Hichme M., Wegenek W. (1998) Toxicol. Appl. Pharmacol. 152, 276–281 - PubMed
-
- Li H., Poulos T. L. (1997) Nat. Struct. Biol. 4, 140–146 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Chemical Information
Research Materials
Miscellaneous

