The multifunctional protein in peroxisomal beta-oxidation: structure and substrate specificity of the Arabidopsis thaliana protein MFP2
- PMID: 20463021
- PMCID: PMC2911295
- DOI: 10.1074/jbc.M110.106005
The multifunctional protein in peroxisomal beta-oxidation: structure and substrate specificity of the Arabidopsis thaliana protein MFP2
Abstract
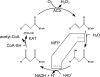
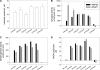
Plant fatty acids can be completely degraded within the peroxisomes. Fatty acid degradation plays a role in several plant processes including plant hormone synthesis and seed germination. Two multifunctional peroxisomal isozymes, MFP2 and AIM1, both with 2-trans-enoyl-CoA hydratase and l-3-hydroxyacyl-CoA dehydrogenase activities, function in mouse ear cress (Arabidopsis thaliana) peroxisomal beta-oxidation, where fatty acids are degraded by the sequential removal of two carbon units. A deficiency in either of the two isozymes gives rise to a different phenotype; the biochemical and molecular background for these differences is not known. Structure determination of Arabidopsis MFP2 revealed that plant peroxisomal MFPs can be grouped into two families, as defined by a specific pattern of amino acid residues in the flexible loop of the acyl-binding pocket of the 2-trans-enoyl-CoA hydratase domain. This could explain the differences in substrate preferences and specific biological functions of the two isozymes. The in vitro substrate preference profiles illustrate that the Arabidopsis AIM1 hydratase has a preference for short chain acyl-CoAs compared with the Arabidopsis MFP2 hydratase. Remarkably, neither of the two was able to catabolize enoyl-CoA substrates longer than 14 carbon atoms efficiently, suggesting the existence of an uncharacterized long chain enoyl-CoA hydratase in Arabidopsis peroxisomes.
Figures


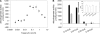


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

