Peroxisomal plant 3-ketoacyl-CoA thiolase structure and activity are regulated by a sensitive redox switch
- PMID: 20463027
- PMCID: PMC2911321
- DOI: 10.1074/jbc.M110.106013
Peroxisomal plant 3-ketoacyl-CoA thiolase structure and activity are regulated by a sensitive redox switch
Abstract
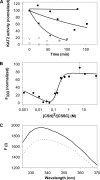
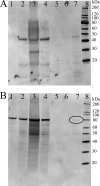
The breakdown of fatty acids, performed by the beta-oxidation cycle, is crucial for plant germination and sustainability. beta-Oxidation involves four enzymatic reactions. The final step, in which a two-carbon unit is cleaved from the fatty acid, is performed by a 3-ketoacyl-CoA thiolase (KAT). The shortened fatty acid may then pass through the cycle again (until reaching acetoacetyl-CoA) or be directed to a different cellular function. Crystal structures of KAT from Arabidopsis thaliana and Helianthus annuus have been solved to 1.5 and 1.8 A resolution, respectively. Their dimeric structures are very similar and exhibit a typical thiolase-like fold; dimer formation and active site conformation appear in an open, active, reduced state. Using an interdisciplinary approach, we confirmed the potential of plant KATs to be regulated by the redox environment in the peroxisome within a physiological range. In addition, co-immunoprecipitation studies suggest an interaction between KAT and the multifunctional protein that is responsible for the preceding two steps in beta-oxidation, which would allow a route for substrate channeling. We suggest a model for this complex based on the bacterial system.
Figures



Similar articles
-
The crystal structure of a plant 3-ketoacyl-CoA thiolase reveals the potential for redox control of peroxisomal fatty acid beta-oxidation.J Mol Biol. 2006 Jun 2;359(2):347-57. doi: 10.1016/j.jmb.2006.03.032. Epub 2006 Mar 29. J Mol Biol. 2006. PMID: 16630629
-
Requirement for 3-ketoacyl-CoA thiolase-2 in peroxisome development, fatty acid beta-oxidation and breakdown of triacylglycerol in lipid bodies of Arabidopsis seedlings.Plant J. 2001 Oct;28(1):1-12. doi: 10.1046/j.1365-313x.2001.01095.x. Plant J. 2001. PMID: 11696182
-
Knockout of the two evolutionarily conserved peroxisomal 3-ketoacyl-CoA thiolases in Arabidopsis recapitulates the abnormal inflorescence meristem 1 phenotype.J Exp Bot. 2014 Dec;65(22):6723-33. doi: 10.1093/jxb/eru397. Epub 2014 Oct 7. J Exp Bot. 2014. PMID: 25297549 Free PMC article.
-
New insights in peroxisomal beta-oxidation. Implications for human peroxisomal disorders.Verh K Acad Geneeskd Belg. 1998;60(3):195-214. Verh K Acad Geneeskd Belg. 1998. PMID: 9803880 Review.
-
Metabolic aspects of peroxisomal beta-oxidation.Biochim Biophys Acta. 1991 Sep 11;1085(2):141-58. doi: 10.1016/0005-2760(91)90089-z. Biochim Biophys Acta. 1991. PMID: 1892883 Review.
Cited by
-
Machine learning-aided microRNA discovery for olive oil quality.PLoS One. 2024 Oct 11;19(10):e0311569. doi: 10.1371/journal.pone.0311569. eCollection 2024. PLoS One. 2024. PMID: 39392838 Free PMC article.
-
Interactome of Arabidopsis Thaliana.Plants (Basel). 2022 Jan 27;11(3):350. doi: 10.3390/plants11030350. Plants (Basel). 2022. PMID: 35161331 Free PMC article.
-
The 3-ketoacyl-CoA thiolase: an engineered enzyme for carbon chain elongation of chemical compounds.Appl Microbiol Biotechnol. 2020 Oct;104(19):8117-8129. doi: 10.1007/s00253-020-10848-w. Epub 2020 Aug 24. Appl Microbiol Biotechnol. 2020. PMID: 32830293 Review.
-
Catabolism of the Cholesterol Side Chain in Mycobacterium tuberculosis Is Controlled by a Redox-Sensitive Thiol Switch.ACS Infect Dis. 2017 Sep 8;3(9):666-675. doi: 10.1021/acsinfecdis.7b00072. Epub 2017 Aug 16. ACS Infect Dis. 2017. PMID: 28786661 Free PMC article.
-
Defining the plant peroxisomal proteome: from Arabidopsis to rice.Front Plant Sci. 2011 Dec 27;2:103. doi: 10.3389/fpls.2011.00103. eCollection 2011. Front Plant Sci. 2011. PMID: 22645559 Free PMC article.
References
-
- Knoop F. (1905) Beitr. Chem. Physiol. Pathol. 6, 150–162
-
- Kindl H. (1987) in The Biochemistry of Plants, Stumpf P.K., and Vol.9, pp. 31–52, Academic Press, London
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

