A limited number of simian immunodeficiency virus (SIV) env variants are transmitted to rhesus macaques vaginally inoculated with SIVmac251
- PMID: 20463069
- PMCID: PMC2898254
- DOI: 10.1128/JVI.00481-10
A limited number of simian immunodeficiency virus (SIV) env variants are transmitted to rhesus macaques vaginally inoculated with SIVmac251
Abstract
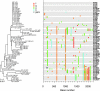
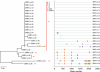
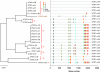
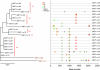
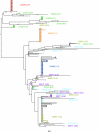
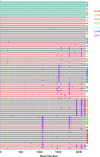
Single-genome amplification (SGA) and sequencing of HIV-1 RNA in plasma of acutely infected humans allows the identification and enumeration of transmitted/founder viruses responsible for productive systemic infection. Use of this strategy as a means for identifying transmitted viruses suggested that intrarectal simian immunodeficiency virus (SIV) inoculation of macaques recapitulates key features of human rectal infection. However, no studies have used the SGA strategy to identify vaginally transmitted virus(es) in macaques or to determine how early SIV diversification in vaginally infected animals compares with HIV-1 in humans. We used SGA to amplify 227 partial env sequences from a SIVmac251 challenge stock and from seven rhesus macaques at the earliest plasma viral RNA-positive time point after low- and high-dose intravaginal inoculation. Sequences were analyzed phylogenetically to determine the relationship of transmitted/founder viruses within and between each animal and the challenge stock. In each animal, discrete low-diversity env sequence lineages were evident, and these coalesced phylogenetically to identical or near-identical env sequences in the challenge stock, thus confirming the validity of the SGA sequencing and modeling strategy for identifying vaginally transmitted SIV. Between 1 and 10 viruses were responsible for systemic infection, similar to humans infected by sexual contact, and the set of viruses transmitted to the seven animals studied represented the full genetic constellation of the challenge stock. These findings recapitulate many of the features of sexual HIV-1 transmission in women. Furthermore, the SIV rhesus macaque model can be used to understand the factors that influence the transmission of single versus multiple SIV variants.
Figures



References
-
- Abrahams, M. R., J. A. Anderson, E. E. Giorgi, C. Seoighe, K. Mlisana, L. H. Ping, G. S. Athreya, F. K. Treurnicht, B. F. Keele, N. Wood, J. F. Salazar-Gonzalez, T. Bhattacharya, H. Chu, I. Hoffman, S. Galvin, C. Mapanje, P. Kazembe, R. Thebus, S. Fiscus, W. Hide, M. S. Cohen, S. A. Karim, B. F. Haynes, G. M. Shaw, B. H. Hahn, B. T. Korber, R. Swanstrom, and C. Williamson. 2009. Quantitating the multiplicity of infection with human immunodeficiency virus type 1 subtype C reveals a non-Poisson distribution of transmitted variants. J. Virol. 83:3556-3567. - PMC - PubMed
-
- Clamp, M., J. Cuff, S. M. Searle, and G. J. Barton. 2004. The Jalview Java alignment editor. Bioinformatics 20:426-427. - PubMed
-
- Felsenstein, J. 1988. Phylogenies from molecular sequences: inference and reliability. Annu. Rev. Genet. 22:521-565. - PubMed
-
- Greenier, J. L., C. J. Miller, D. Lu, P. J. Dailey, F. X. Lu, K. J. Kunstman, S. M. Wolinsky, and M. L. Marthas. 2001. Route of simian immunodeficiency virus inoculation determines the complexity but not the identity of viral variant populations that infect rhesus macaques. J. Virol. 75:3753-3765. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

