Nicotinamide adenine dinucleotide-dependent binding of the neuronal Ca2+ sensor protein GCAP2 to photoreceptor synaptic ribbons
- PMID: 20463219
- PMCID: PMC3900572
- DOI: 10.1523/JNEUROSCI.3701-09.2010
Nicotinamide adenine dinucleotide-dependent binding of the neuronal Ca2+ sensor protein GCAP2 to photoreceptor synaptic ribbons
Abstract
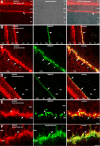
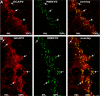
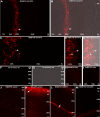
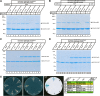
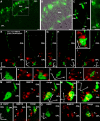
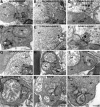
Guanylate cyclase activating protein 2 (GCAP2) is a recoverin-like Ca2+-sensor protein known to modulate guanylate cyclase activity in photoreceptor outer segments. GCAP2 is also present in photoreceptor ribbon synapses where its function is unknown. Synaptic ribbons are active zone-associated presynaptic structures in the tonically active photoreceptor ribbon synapses and contain RIBEYE as a unique and major protein component. In the present study, we demonstrate by various independent approaches that GCAP2 specifically interacts with RIBEYE in photoreceptor synapses. We show that the flexible hinge 2 linker region of RIBEYE(B) domain that connects the nicotinamide adenine dinucleotide (NADH)-binding subdomain with the substrate-binding subdomain (SBD) binds to the C terminus of GCAP2. We demonstrate that the RIBEYE-GCAP2 interaction is induced by the binding of NADH to RIBEYE. RIBEYE-GCAP2 interaction is modulated by the SBD. GCAP2 is strongly expressed in synaptic terminals of light-adapted photoreceptors where GCAP2 is found close to synaptic ribbons as judged by confocal microscopy and proximity ligation assays. Virus-mediated overexpression of GCAP2 in photoreceptor synaptic terminals leads to a reduction in the number of synaptic ribbons. Therefore, GCAP2 is a prime candidate for mediating Ca2+-dependent dynamic changes of synaptic ribbons in photoreceptor synapses.
Figures





References
-
- Ames JB, Dizhoor AM, Ikura M, Palczewski K, Stryer L. Three-dimensional structure of guanylyl cyclase activating protein-2, a calcium-sensitive modulator of photoreceptor guanylyl cyclases. J Biol Chem. 1999;274:19329–19337. - PubMed
-
- Ashery U, Betz A, Xu T, Brose N, Rettig J. An efficient method for the infection of adrenal chromaffin cells using the Semliki Forest virus gene expression system. Eur J Cell Biol. 1999;78:525–532. - PubMed
-
- Balasubramanian P, Zhao LJ, Chinnadurai G. Nicotinamide adenine dinucleotide stimulates oligomerization, interaction with E1A and an intrinsic dehydrogenase activity of CtBP. FEBS Lett. 2003;537:157–160. - PubMed
-
- Barnes CJ, Vadlamudi RK, Mishra SK, Jacobson RH, Liu F, Kumar R. Functional inactivation of a transcriptional corepressor by a signalling kinase. Nat Struct Biol. 2003;10:622–628. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
