S-Nitrosylation of DRP1 does not affect enzymatic activity and is not specific to Alzheimer's disease
- PMID: 20463395
- PMCID: PMC2893334
- DOI: 10.3233/JAD-2010-100552
S-Nitrosylation of DRP1 does not affect enzymatic activity and is not specific to Alzheimer's disease
Abstract
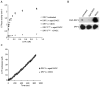
Mitochondrial dysfunction and synaptic loss are among the earliest events linked to Alzheimer's disease (AD) and might play a causative role in disease onset and progression. The underlying mechanisms of mitochondrial and synaptic dysfunction in AD remain unclear. We previously reported that nitric oxide (NO) triggers persistent mitochondrial fission and causes neuronal cell death. A recent article claimed that S-nitrosylation of dynamin related protein 1 (DRP1) at cysteine 644 causes protein dimerization and increased GTPase activity and is the mechanism responsible for NO-induced mitochondrial fission and neuronal injury in AD, but not in Parkinson's disease (PD). However, this report remains controversial. To resolve the controversy, we investigated the effects of S-nitrosylation on DRP1 structure and function. Contrary to the previous report, S-nitrosylation of DRP1 does not increase GTPase activity or cause dimerization. In fact, DRP1 does not exist as a dimer under native conditions, but rather as a tetramer capable of self-assembly into higher order spiral- and ring-like oligomeric structures after nucleotide binding. S-nitrosylation, as confirmed by the biotin-switch assay, has no impact on DRP1 oligomerization. Importantly, we found no significant difference in S-nitrosylated DRP1 (SNO-DRP1) levels in brains of age-matched normal, AD, or PD patients. We also found that S-nitrosylation is not specific to DRP1 because S-nitrosylated optic atrophy 1 (SNO-OPA1) is present at comparable levels in all human brain samples. Finally, we show that NO triggers DRP1 phosphorylation at serine 616, which results in its activation and recruitment to mitochondria. Our data indicate the mechanism underlying nitrosative stress-induced mitochondrial fragmentation in AD is not DRP1 S-nitrosylation.
Figures



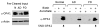

References
-
- Lu T, Pan Y, Kao SY, Li C, Kohane I, Chan J, Yankner BA. Gene regulation and DNA damage in the ageing human brain. Nature. 2004;429:883–891. - PubMed
-
- Nunomura A, Perry G, Aliev G, Hirai K, Takeda A, Balraj EK, Jones PK, Ghanbari H, Wataya T, Shimohama S, Chiba S, Atwood CS, Petersen RB, Smith MA. Oxidative damage is the earliest event in Alzheimer disease. J Neuropathol Exp Neurol. 2001;60:759–767. - PubMed
-
- Tamagno E, Parola M, Bardini P, Piccini A, Borghi R, Guglielmotto M, Santoro G, Davit A, Danni O, Smith MA, Perry G, Tabaton M. Beta-site APP cleaving enzyme up-regulation induced by 4-hydroxynonenal is mediated by stress-activated protein kinases pathways. J Neurochem. 2005;92:628–636. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

