A Bayesian framework to account for complex non-genetic factors in gene expression levels greatly increases power in eQTL studies
- PMID: 20463871
- PMCID: PMC2865505
- DOI: 10.1371/journal.pcbi.1000770
A Bayesian framework to account for complex non-genetic factors in gene expression levels greatly increases power in eQTL studies
Abstract
Gene expression measurements are influenced by a wide range of factors, such as the state of the cell, experimental conditions and variants in the sequence of regulatory regions. To understand the effect of a variable of interest, such as the genotype of a locus, it is important to account for variation that is due to confounding causes. Here, we present VBQTL, a probabilistic approach for mapping expression quantitative trait loci (eQTLs) that jointly models contributions from genotype as well as known and hidden confounding factors. VBQTL is implemented within an efficient and flexible inference framework, making it fast and tractable on large-scale problems. We compare the performance of VBQTL with alternative methods for dealing with confounding variability on eQTL mapping datasets from simulations, yeast, mouse, and human. Employing Bayesian complexity control and joint modelling is shown to result in more precise estimates of the contribution of different confounding factors resulting in additional associations to measured transcript levels compared to alternative approaches. We present a threefold larger collection of cis eQTLs than previously found in a whole-genome eQTL scan of an outbred human population. Altogether, 27% of the tested probes show a significant genetic association in cis, and we validate that the additional eQTLs are likely to be real by replicating them in different sets of individuals. Our method is the next step in the analysis of high-dimensional phenotype data, and its application has revealed insights into genetic regulation of gene expression by demonstrating more abundant cis-acting eQTLs in human than previously shown. Our software is freely available online at http://www.sanger.ac.uk/resources/software/peer/.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

 matrix
matrix  of measured gene expression levels of
of measured gene expression levels of  genes from
genes from  individuals is modelled by additive contributions from components
individuals is modelled by additive contributions from components  and observation noise
and observation noise  . Here, the components capture the signal due to primary effect of the genetic state
. Here, the components capture the signal due to primary effect of the genetic state  , known factors
, known factors  and hidden factors
and hidden factors  . Some examples of possible underlying sources of variation are given above the model boxes. The groupings represent some standard genetic association models commonly used.
. Some examples of possible underlying sources of variation are given above the model boxes. The groupings represent some standard genetic association models commonly used.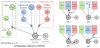
 . The solid rectangles indicate that contained variables are duplicated for each gene probe (
. The solid rectangles indicate that contained variables are duplicated for each gene probe ( ), SNP (
), SNP ( ) or factor (
) or factor ( ) respectively. A similar rectangle for individuals (
) respectively. A similar rectangle for individuals ( ) is omitted in this representation. The dashed rectangle indicates that the variable
) is omitted in this representation. The dashed rectangle indicates that the variable  switches the contained part of the graph on or off representing the existence or lack of an association. Nodes with thick outlines (
switches the contained part of the graph on or off representing the existence or lack of an association. Nodes with thick outlines ( ,
,  and
and  ) are observed. (b)–(e) Update cycle of the known factors model introduced in Section Inference. The red outline highlights the parts of the model that change in a step, and the thick blue arrows illustrate the flow of information. Details of these updates are discussed in the text.
) are observed. (b)–(e) Update cycle of the known factors model introduced in Section Inference. The red outline highlights the parts of the model that change in a step, and the thick blue arrows illustrate the flow of information. Details of these updates are discussed in the text.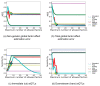
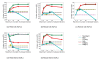
 ), thus only a single count is given (dashed lines), together with the number of factors found (diamond shape). Other methods were applied with a maximum number of
), thus only a single count is given (dashed lines), together with the number of factors found (diamond shape). Other methods were applied with a maximum number of  ,
,  ,
,  and
and  hidden factors.
hidden factors.

References
-
- Brem RB, Yvert G, Clinton R, Kruglyak L. Genetic dissection of transcriptional regulation in budding yeast. Science. 2002;296:752–755. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

