Modeling reveals bistability and low-pass filtering in the network module determining blood stem cell fate
- PMID: 20463872
- PMCID: PMC2865510
- DOI: 10.1371/journal.pcbi.1000771
Modeling reveals bistability and low-pass filtering in the network module determining blood stem cell fate
Abstract
Combinatorial regulation of gene expression is ubiquitous in eukaryotes with multiple inputs converging on regulatory control elements. The dynamic properties of these elements determine the functionality of genetic networks regulating differentiation and development. Here we propose a method to quantitatively characterize the regulatory output of distant enhancers with a biophysical approach that recursively determines free energies of protein-protein and protein-DNA interactions from experimental analysis of transcriptional reporter libraries. We apply this method to model the Scl-Gata2-Fli1 triad-a network module important for cell fate specification of hematopoietic stem cells. We show that this triad module is inherently bistable with irreversible transitions in response to physiologically relevant signals such as Notch, Bmp4 and Gata1 and we use the model to predict the sensitivity of the network to mutations. We also show that the triad acts as a low-pass filter by switching between steady states only in response to signals that persist for longer than a minimum duration threshold. We have found that the auto-regulation loops connecting the slow-degrading Scl to Gata2 and Fli1 are crucial for this low-pass filtering property. Taken together our analysis not only reveals new insights into hematopoietic stem cell regulatory network functionality but also provides a novel and widely applicable strategy to incorporate experimental measurements into dynamical network models.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
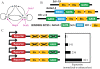

 and
and  . In B and C, we use different values for these chromatin equilibrium constants and recalculate all free energy values using the analytical equations derived with experimental results. For
. In B and C, we use different values for these chromatin equilibrium constants and recalculate all free energy values using the analytical equations derived with experimental results. For  in B, only Bmp4 can switch the triad from OFF to ON. For
in B, only Bmp4 can switch the triad from OFF to ON. For  (C) neither Notch nor Bmp4 can switch the triad to ON state. D. Bistable response of the triad module to Gata1 repressor signal. Gata1 competes with Gata2 for binding sites on the Gata2-3 enhancer and can switch the triad from ON state to OFF by decreasing the recruitment of RNA polymerase to the Gata2 promoter by a factor
(C) neither Notch nor Bmp4 can switch the triad to ON state. D. Bistable response of the triad module to Gata1 repressor signal. Gata1 competes with Gata2 for binding sites on the Gata2-3 enhancer and can switch the triad from ON state to OFF by decreasing the recruitment of RNA polymerase to the Gata2 promoter by a factor  . As a result the system irreversibly switches from ON to OFF. (note that this figure is shown in linear scale, the inset shows the deactivation in log-log scale for comparison with A). To evaluate the steady state dose response of each signal individually the concentrations of other signals were kept fixed at zero during simulation.
. As a result the system irreversibly switches from ON to OFF. (note that this figure is shown in linear scale, the inset shows the deactivation in log-log scale for comparison with A). To evaluate the steady state dose response of each signal individually the concentrations of other signals were kept fixed at zero during simulation.

Similar articles
-
Gata2, Fli1, and Scl form a recursively wired gene-regulatory circuit during early hematopoietic development.Proc Natl Acad Sci U S A. 2007 Nov 6;104(45):17692-7. doi: 10.1073/pnas.0707045104. Epub 2007 Oct 25. Proc Natl Acad Sci U S A. 2007. PMID: 17962413 Free PMC article.
-
Mathematical model of a gene regulatory network reconciles effects of genetic perturbations on hematopoietic stem cell emergence.Dev Biol. 2013 Jul 15;379(2):258-69. doi: 10.1016/j.ydbio.2013.04.016. Epub 2013 Apr 23. Dev Biol. 2013. PMID: 23623899
-
A regulatory network governing Gata1 and Gata2 gene transcription orchestrates erythroid lineage differentiation.Int J Hematol. 2014 Nov;100(5):417-24. doi: 10.1007/s12185-014-1568-0. Epub 2014 Mar 18. Int J Hematol. 2014. PMID: 24638828 Review.
-
Thermodynamic models of combinatorial gene regulation by distant enhancers.IET Syst Biol. 2010 Nov;4(6):393-408. doi: 10.1049/iet-syb.2010.0010. IET Syst Biol. 2010. PMID: 21073238
-
SCL/TAL1 in Hematopoiesis and Cellular Reprogramming.Curr Top Dev Biol. 2016;118:163-204. doi: 10.1016/bs.ctdb.2016.01.004. Epub 2016 Feb 18. Curr Top Dev Biol. 2016. PMID: 27137657 Review.
Cited by
-
Assessing regulatory information in developmental gene regulatory networks.Proc Natl Acad Sci U S A. 2017 Jun 6;114(23):5862-5869. doi: 10.1073/pnas.1610616114. Proc Natl Acad Sci U S A. 2017. PMID: 28584110 Free PMC article.
-
Emerging properties of animal gene regulatory networks.Nature. 2010 Dec 16;468(7326):911-20. doi: 10.1038/nature09645. Nature. 2010. PMID: 21164479 Free PMC article. Review.
-
The transcriptional programme controlled by Runx1 during early embryonic blood development.Dev Biol. 2012 Jun 15;366(2):404-19. doi: 10.1016/j.ydbio.2012.03.024. Epub 2012 Apr 21. Dev Biol. 2012. PMID: 22554697 Free PMC article.
-
How Does the Regulatory Genome Work?J Comput Biol. 2019 Jul;26(7):685-695. doi: 10.1089/cmb.2019.0097. Epub 2019 Jun 4. J Comput Biol. 2019. PMID: 31166788 Free PMC article.
-
Kinetic models of hematopoietic differentiation.Wiley Interdiscip Rev Syst Biol Med. 2019 Jan;11(1):e1424. doi: 10.1002/wsbm.1424. Epub 2018 Apr 16. Wiley Interdiscip Rev Syst Biol Med. 2019. PMID: 29660842 Free PMC article. Review.
References
-
- Davidson EH. The regulatory genome : gene regulatory networks in development and evolution. Amsterdam ; Boston: Academic Press.; 2006. p. xi, 289.
-
- Chaves M, Albert R, Sontag ED. Robustness and fragility of Boolean models for genetic regulatory networks. J Theor Biol. 2005;235(3):431–49. - PubMed
-
- Laslo P, Spooner CJ, Warmflash A, Lancki DW, Lee HJ, et al. Multilineage transcriptional priming and determination of alternate hematopoietic cell fates. Cell. 2006;126(4):755–66. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

