Linkage and association mapping of Arabidopsis thaliana flowering time in nature
- PMID: 20463887
- PMCID: PMC2865524
- DOI: 10.1371/journal.pgen.1000940
Linkage and association mapping of Arabidopsis thaliana flowering time in nature
Abstract
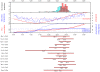
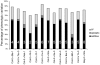
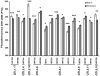
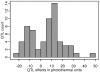
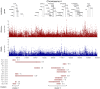
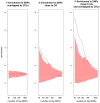
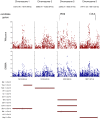
Flowering time is a key life-history trait in the plant life cycle. Most studies to unravel the genetics of flowering time in Arabidopsis thaliana have been performed under greenhouse conditions. Here, we describe a study about the genetics of flowering time that differs from previous studies in two important ways: first, we measure flowering time in a more complex and ecologically realistic environment; and, second, we combine the advantages of genome-wide association (GWA) and traditional linkage (QTL) mapping. Our experiments involved phenotyping nearly 20,000 plants over 2 winters under field conditions, including 184 worldwide natural accessions genotyped for 216,509 SNPs and 4,366 RILs derived from 13 independent crosses chosen to maximize genetic and phenotypic diversity. Based on a photothermal time model, the flowering time variation scored in our field experiment was poorly correlated with the flowering time variation previously obtained under greenhouse conditions, reinforcing previous demonstrations of the importance of genotype by environment interactions in A. thaliana and the need to study adaptive variation under natural conditions. The use of 4,366 RILs provides great power for dissecting the genetic architecture of flowering time in A. thaliana under our specific field conditions. We describe more than 60 additive QTLs, all with relatively small to medium effects and organized in 5 major clusters. We show that QTL mapping increases our power to distinguish true from false associations in GWA mapping. QTL mapping also permits the identification of false negatives, that is, causative SNPs that are lost when applying GWA methods that control for population structure. Major genes underpinning flowering time in the greenhouse were not associated with flowering time in this study. Instead, we found a prevalence of genes involved in the regulation of the plant circadian clock. Furthermore, we identified new genomic regions lacking obvious candidate genes.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
References
-
- Jones H, Leigh FJ, Mackay I, Bower MA, Smith LMJ, et al. Population-based resequencing reveals that the flowering time adaptation of cultivated barley originated east of the Fertile Crescent. Mol Biol Evol. 2008;25:2211–2219. - PubMed
-
- Uga Y, Nonoue Y, Liang Z, Lin H, Yamamoto S, et al. Accumulation of additive effects generates a strong photoperiod sensitivity in the extremely late-heading rice cultivar ‘Nona Bokra’. Theor Appl Genet. 2007;114:1457–1466. - PubMed
-
- Van Dijk H, Hautekèete N-C. Long day plants and the response to global warming: rapid evolutionary change in day length sensitivity is possible in wild beet. J Evol Biol. 2007;20:349–357. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

